The overuse of antibiotics in the aquaculture industry has led to the emergence of antimicrobial resistance, a significant threat to public health and the environment. In this context, bacteriophage (phage) therapy—which uses viruses that specifically infect and destroy bacteria—is emerging as a promising and sustainable alternative.
However, the efficacy of phage therapy is not guaranteed, as bacteria have evolved a sophisticated arsenal of defense mechanisms to protect themselves from phage attacks. Understanding this protective “armor” is critical for designing effective treatments. A recent review study published in Aquaculture by researchers from Tezpur University, Koneru Lakshmaiah Education Foundation, and Rabindranath Tagore University has comprehensively mapped the landscape of these defense systems in the most relevant bacterial pathogens in aquaculture, offering a crucial guide for the future of aquatic animal health.
- 1 Key findings
- 2 Phage therapy in aquaculture: Promise and challenges
- 3 A genomic arsenal: Over 1,100 defense systems identified
- 4 The most common defense mechanisms in fish pathogens
- 5 Defensive strategies vary among major pathogens
- 6 Implications for smarter, more robust phage therapy
- 7 Entradas relacionadas:
Key findings
- A total of 1,174 anti-phage defense systems were identified across the genomes of 272 bacterial strains, covering 35 pathogenic species significant to aquaculture.
- While “classic” systems, such as Restriction-Modification (R-M) and CRISPR-Cas, are common, a vast diversity of emerging mechanisms with lineage-specific distributions also exists.
- Defenses are often clustered in “genomic islands,” suggesting they are acquired modularly and can be transferred horizontally between bacteria, facilitating rapid adaptation.
- Pathogens such as Vibrio and Aeromonas rely more on innate systems (R-M, abortive infection), whereas Flavobacterium actively uses adaptive CRISPR-Cas immunity, indicating direct co-evolution with phages in farming environments.
- Knowledge of a pathogen’s defensive repertoire is crucial for designing more effective phage therapies, such as rotating phage cocktails or using engineered phages to overcome these barriers.
Phage therapy in aquaculture: Promise and challenges
Phage therapy leverages the lytic action of these viruses to eliminate pathogenic bacteria with high specificity, leaving the beneficial environmental microbiota unharmed. Since its first reported success in 1981 against Aeromonas hydrophila, numerous laboratory studies have demonstrated its potential to reduce pathogen loads and improve fish survival rates.
Nevertheless, transitioning from the laboratory to real-world aquaculture farm conditions presents significant challenges. Environmental factors such as temperature, pH, salinity, and organic load can impact phage stability and efficacy. Furthermore, bacteria can develop resistance, necessitating more complex strategies like “phage cocktails” that target the pathogen from multiple angles. The primary obstacle, however, lies within the bacterial genome itself: a vast repertoire of defense systems ready for activation.
A genomic arsenal: Over 1,100 defense systems identified
To grasp the scale of this arsenal, researchers analyzed 272 high-quality genomes representing 35 species of fish-pathogenic bacteria. Using bioinformatic tools, they identified a total of 1,174 anti-phage defense systems, classified into 304 distinct types.
These systems are not randomly distributed. They are frequently found clustered in specific genomic regions known as “defense islands.” This organization suggests that the systems can be acquired and shared among bacteria via horizontal gene transfer, enabling swift adaptation to phage pressure in aquatic environments.
The most common defense mechanisms in fish pathogens
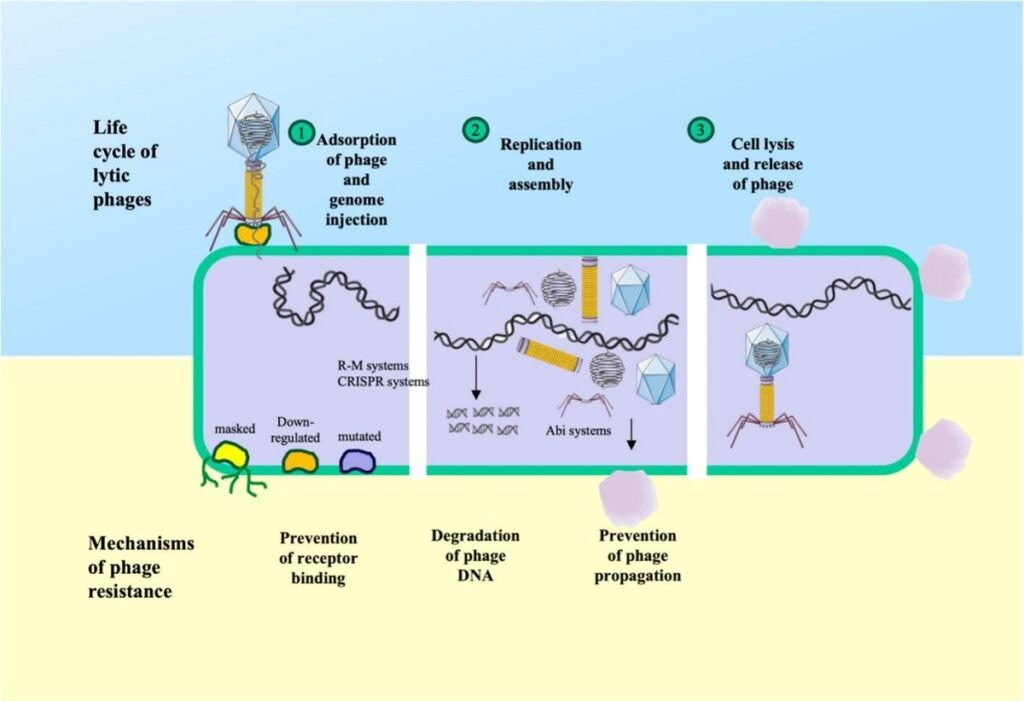
While immensely diverse, most defenses can be grouped into several core strategies that bacteria employ to survive a phage infection.
Restriction-modification (R-M) systems: The first line of defense
This is one of the most widespread systems, functioning as a “self vs. non-self” discrimination mechanism. A methyltransferase enzyme marks the bacterium’s own DNA with a chemical tag (methylation). When a phage injects its DNA, a restriction endonuclease enzyme inspects it. If the phage DNA lacks the correct tag, it is identified as foreign and destroyed. R-M systems have been found in key pathogens like Aeromonas hydrophila, Vibrio anguillarum, and Piscirickettsia salmonis.
CRISPR-Cas: Adaptive immune memory
The CRISPR-Cas system is more sophisticated, providing heritable adaptive immunity. When a bacterium is first attacked by a phage, it can capture a small piece of the phage’s DNA and store it in a region of its own genome called the CRISPR locus. If the same or a similar phage attacks again, the bacterium uses this stored fragment as a guide to recognize and precisely destroy the invader’s genetic material. This mechanism is particularly prevalent and active in species like Flavobacterium columnare, where a constant co-evolution between bacterial CRISPR defenses and phage mutations has been observed on fish farms.
Stay Always Informed
Join our communities to instantly receive the most important news, reports, and analysis from the aquaculture industry.
Abortive infection (Abi): Sacrifice for the greater good
If the previous systems fail, some bacteria resort to an altruistic strategy of cellular suicide. Abortive infection (Abi) systems activate upon infection and either halt the cell’s metabolism or destroy it entirely before the phage can complete its replication cycle and produce new viral particles. Although the infected cell dies, this sacrifice prevents the phage from spreading and protects the rest of the bacterial population. Abi systems have been identified in pathogens such as Aeromonas salmonicida and Flavobacterium spp.
Defensive strategies vary among major pathogens
The study reveals that different pathogens rely on distinct defensive strategies.
- Vibrio and Aeromonas: These genera, responsible for diseases like vibriosis and hemorrhagic septicemia, show a low prevalence of CRISPR-Cas systems. Instead, they primarily depend on R-M systems, surface-level defenses (like receptor modification), and abortive infection systems.
- Flavobacterium: Unlike the others, pathogens such as F. columnare and F. psychrophilum actively use CRISPR-Cas systems as a dynamic defense, continuously acquiring new phage sequences to adapt to local threats.
- Piscirickettsia salmonis: This intracellular pathogen, which causes salmonid rickettsial septicaemia, features a highly complex, multi-layered defense repertoire. Genomic analysis has revealed systems for depleting phage DNA building blocks (dGTPase), abortive infection systems (AbiD), and multiple toxin-antitoxin systems, many located on plasmids, which facilitates their dissemination.
Implications for smarter, more robust phage therapy
The main conclusion of this comprehensive review is clear: for phage therapy to become a reliable and durable tool in aquaculture, it is essential to understand the enemy and its defenses. Ignoring the bacterial “armor” can lead to treatment failure.
The study proposes a pragmatic roadmap:
- Profile defenses: Before applying treatment, analyze the defense repertoire of the target pathogen.
- Design strategic cocktails: Use combinations of phages that bacteria cannot easily counter, or rotate cocktails to prevent the emergence of widespread resistance.
- Develop “Enhanced” phages: Genetic engineering can create phages capable of evading specific defenses, for instance, by producing proteins (anti-CRISPRs) that inactivate bacterial CRISPR systems.
With a more rational, evidence-based approach, phage therapy can transition from a promising concept to a robust modality aligned with sustainable management, reducing antibiotic dependency and strengthening the long-term resilience of global aquaculture.
Contact
Aditya Kumar
Department of Molecular Biology and Biotechnology, Tezpur University, Tezpur 784028, Assam, India
Email: aditya@tezu.ernet.in
Reference
Olymon, K., Kinoo, N., Roy, N., Yella, V. R., Teronpi, V., & Kumar, A. (2025). Fish pathogen armor: A review of antiphage defense systems in bacterial fish pathogens. Aquaculture, 743192. https://doi.org/10.1016/j.aquaculture.2025.743192
Editor at the digital magazine AquaHoy. He holds a degree in Aquaculture Biology from the National University of Santa (UNS) and a Master’s degree in Science and Innovation Management from the Polytechnic University of Valencia, with postgraduate diplomas in Business Innovation and Innovation Management. He possesses extensive experience in the aquaculture and fisheries sector, having led the Fisheries Innovation Unit of the National Program for Innovation in Fisheries and Aquaculture (PNIPA). He has served as a senior consultant in technology watch, an innovation project formulator and advisor, and a lecturer at UNS. He is a member of the Peruvian College of Biologists and was recognized by the World Aquaculture Society (WAS) in 2016 for his contribution to aquaculture.