Recirculating Aquaculture Systems (RAS) are cornerstones of sustainable aquaculture, yet their high stocking densities make them vulnerable to rapidly spreading disease outbreaks. While identifying the causative agent promptly is crucial, traditional methods like culturing and PCR often “mask” the true bacterial diversity and pathogen prevalence.
Motivated by a mortality event at a commercial Atlantic salmon (Salmo salar) RAS facility in Chile, a recent study employed Oxford Nanopore Technology (ONT) sequencing to obtain a high-definition microbial profile and uncover the culprit. The research was published by investigators from the Interdisciplinary Center for Aquaculture Research (INCAR) at the University of Concepción, Universidad San Sebastián, and MOWI Chile SA. The findings reveal not only the responsible pathogen but also the functional differences in the gut microbiota that separate a healthy fish from a diseased one.
- 1 Key findings
- 2 The challenge: An outbreak of unknown origin
- 3 A new tool: Full-length 16S sequencing
- 4 Key findings: The microbiota of sick vs. healthy salmon
- 5 Beyond who is there: What the bacteria “do”
- 6 The system’s role: Biofilters as reservoirs
- 7 Conclusion: A proactive tool for RAS health
- 8 Entradas relacionadas:
Key findings
- Atlantic salmon presenting with abdominal swelling (diseased) exhibited significantly lower intestinal microbial diversity compared to fish without clinical symptoms.
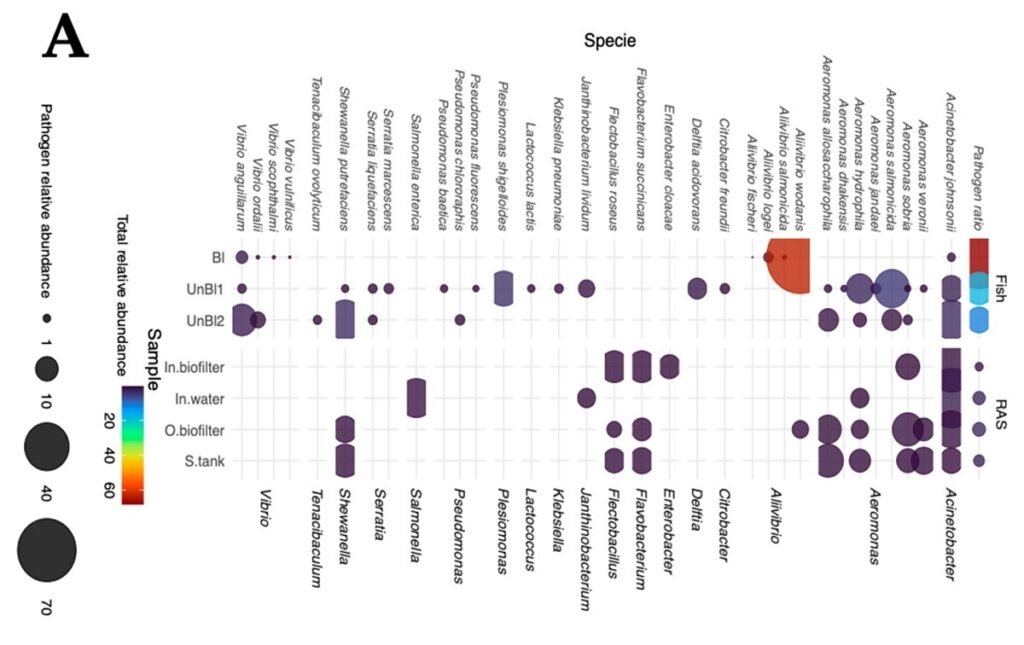
- A high abundance of the pathogen Aliivibrio wodanis was detected in the intestines of the diseased salmon. Notably, this bacterium was absent in the healthy fish.
- Healthy fish showed more metabolic pathways linked to the production of amino acids and short-chain fatty acids, which are crucial for good health. In contrast, diseased fish displayed an increase in sulfur metabolism (H2S production), a function directly associated with the presence of A. wodanis.
- A. wodanis was detected in the RAS biofilter outlet, suggesting that the system’s biofilms may act as reservoirs for this pathogen, enabling cross-contamination between the water and the fish.
- Full-length 16S gene Nanopore sequencing offers greater taxonomic resolution (identification at the species level) and faster results (in ~24h) than traditional PCR or short-read sequencing methods, making it ideal for health surveillance.
The challenge: An outbreak of unknown origin
The study was initiated following a mortality event at a commercial fish farm. The affected salmon showed clear clinical signs: abdominal distension (swelling) and varying degrees of peritonitis and granulomatous inflammation. Facility staff initially suspected known pathogens such as Rhodococcus sp. or Francisella sp., but PCR tests for these agents returned negative. This diagnostic dead end underscored the need for a deeper, broader approach capable of analyzing the entire bacterial community present.
A new tool: Full-length 16S sequencing
Researchers turned to Nanopore-based metagenomics. Unlike common sequencing techniques that only read small variable regions of the 16S rRNA gene (like V3-V4), Nanopore technology allows for sequencing the entire full-length 16S gene (V1-V9).
This “long-read” capability offers much higher taxonomic resolution, enabling more reliable identification of bacteria at the species level.
“One of Nanopore’s advantages is that it allows us to read the entire 16S gene, identifying bacteria down to the species level, which isn’t achieved with other sequencing technologies. Furthermore, it’s fast and portable: in less than a day, we can have a complete overview of the microbial community, including pathogens that traditional methods might overlook,” explained Dr. Diego Valenzuela-Miranda, Adjunct Researcher for INCAR’s “Aquaculture Genomics” line.
For the study, researchers sampled the intestines of diseased (swollen) and healthy (asymptomatic) fish, as well as water samples from key RAS components: the water inlet, the sedimentation tank, and the biofilters.
Key findings: The microbiota of sick vs. healthy salmon
The researchers’ main results revealed the existence of bacteria related to denitrification and hydrogen sulfide metabolism processes in the water. They determined that diseased fish showed lower microbial diversity and a predominance of bacteria from the Proteobacteria group. Furthermore, pathogens such as Aeromonas, Vibrio, and Aliivibrio were detected, including Aliivibrio wodanis in fish with clinical symptoms. In contrast, healthy salmon presented metabolic pathways associated with better digestion and nutrient fermentation.
The sequencing analysis immediately shed light on the fish’s condition. The most evident finding was that diseased fish exhibited drastically reduced intestinal microbial diversity compared to healthy fish—a known indicator of dysbiosis and poor health.
Stay Always Informed
Join our communities to instantly receive the most important news, reports, and analysis from the aquaculture industry.
The dominance of Proteobacteria and a key pathogen
When analyzing the composition, the difference was overwhelming. The intestines of the diseased fish were dominated by over 98% by the phylum Proteobacteria.
Within this group, one pathogen emerged as the primary suspect: Aliivibrio wodanis. This bacterium was found in high concentrations in the fish with abdominal distension, but most revealingly, it was completely absent from the healthy fish samples.
Interestingly, A. wodanis is classically associated with “winter ulcer disease,” but the symptoms observed in this outbreak (swelling and peritonitis) were different. This suggests that the pathogenicity of A. wodanis may vary or manifest differently depending on the system’s conditions.
Opportunistic pathogens: A delicate balance
The study also demonstrated that health is not merely the absence of pathogens. Bacteria from potentially pathogenic genera like Aeromonas and Vibrio (including V. anguillarum) were detected in all fish, both healthy and diseased.
This reinforces the idea that many pathogens are opportunistic. In a healthy, diverse gut, their presence is controlled. However, in a state of dysbiosis (low diversity), these opportunists, or emerging pathogens like A. wodanis, can proliferate and cause disease.
Beyond who is there: What the bacteria “do”
Metagenomics not only identifies who is present but can also predict what metabolic functions the community is performing. Here, the functional differences were as stark as the taxonomic ones.
The functional profile of a healthy fish
The microbiota of healthy fish showed a greater contribution from metabolic pathways related to:
- Amino acid metabolism.
- Short-chain fatty acid (SCFA) fermentation.
Both pathways are fundamental for nutrition, energy metabolism, and the overall well-being of the fish.
The functional profile of a diseased fish
Conversely, the microbiota of diseased fish showed a significant increase in pathways related to sulfur metabolism.
Specifically, this translated into higher production of hydrogen sulfide (H2S). The functional analysis identified A. wodanis as the primary contributor to this H2S production in the diseased fish.
Excess H2S in the gut is problematic; not only is it a toxic compound, but it can also have cytoprotective effects for the bacteria themselves, helping them resist antibiotics and complicating treatment.
The system’s role: Biofilters as reservoirs
The study was not limited to the fish. Analysis of the RAS water revealed that A. wodanis was also detected in the biofilter outlet.
“We detected Aliivibrio wodanis in diseased fish and also in the system’s water, suggesting possible environmental transmission. This demonstrates that biofilters and tanks can act as pathogen reservoirs, making it key to monitor not only the fish but also the microbiological environment of the RAS,” detailed Dr. Valenzuela-Miranda.
This is a critical finding for RAS management. It demonstrates that the biofilms growing on system components (like biofilters) can act as pathogen reservoirs. From there, bacteria can detach, remain in the water column, and cause recolonization or infection of the fish, creating a cycle of cross-contamination.
Conclusion: A proactive tool for RAS health
This study demonstrates the power of full-length 16S Nanopore sequencing as a rapid and precise diagnostic and surveillance tool in aquaculture.
It succeeded where traditional PCR failed: it identified Aliivibrio wodanis as the putatively responsible pathogen for the outbreak, not only due to its presence in diseased fish but also because of its detrimental metabolic function (H2S production).
These findings suggest that using technologies like Nanopore can help monitor microbial health in farming systems more accurately, allowing problems to be detected before they impact production.
“If we incorporate this technology into routine monitoring, we could anticipate outbreaks and act preventively, reducing antibiotic use and improving the health of the fish and the system,” the researcher concluded.
Contact
Diego Valenzuela-Miranda
Interdisciplinary Center for Aquaculture Research (INCAR), University of Concepción
Concepción 4070409, Chile
Email: divalenzuela@udec.cl
Reference (open access)
Valenzuela-Miranda, D., Morales-Rivera, M., Mancilla-Schutz, J., Sandoval, A., Valenzuela-Muñoz, V., & Gallardo-Escárate, C. (2025). Nanopore-Based Metagenomic Approaches for Detection of Bacterial Pathogens in Recirculating Aquaculture Systems. Fishes, 10(10), 496. https://doi.org/10.3390/fishes10100496
Editor at the digital magazine AquaHoy. He holds a degree in Aquaculture Biology from the National University of Santa (UNS) and a Master’s degree in Science and Innovation Management from the Polytechnic University of Valencia, with postgraduate diplomas in Business Innovation and Innovation Management. He possesses extensive experience in the aquaculture and fisheries sector, having led the Fisheries Innovation Unit of the National Program for Innovation in Fisheries and Aquaculture (PNIPA). He has served as a senior consultant in technology watch, an innovation project formulator and advisor, and a lecturer at UNS. He is a member of the Peruvian College of Biologists and was recognized by the World Aquaculture Society (WAS) in 2016 for his contribution to aquaculture.