The aquaculture industry has experienced significant growth in recent years, driven by the increasing global demand for seafood. However, the development of sustainable and efficient aquaculture practices remains a critical challenge.
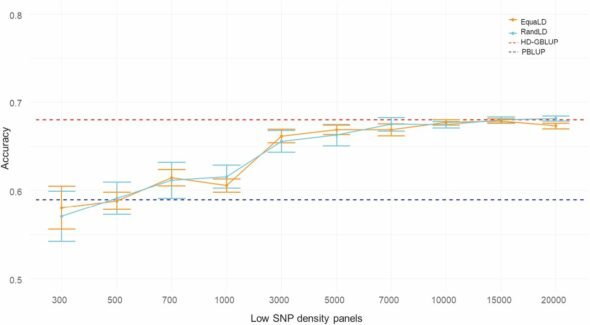
Genomic selection (GS) and marker-assisted selection (MAS) have emerged as powerful tools to accelerate genetic improvement in aquaculture species. However, the cost of high-throughput SNP genotyping can be prohibitive, particularly for species with large breeding populations, such as shrimp.
A team of scientists from the Institute of Oceanology, the University of Chinese Academy of Sciences, and the Qingdao Marine Science and Technology Center employed a bulked segregant analysis sequencing (BSA-seq) method to identify single nucleotide polymorphisms (SNPs) and growth-related genes in the Pacific white shrimp Litopenaeus vannamei.
BSA-seq: A Cost-Effective Alternative
Bulked segregant analysis sequencing (BSA-seq) offers a promising alternative to traditional SNP genotyping methods.
BSA involves combining DNA samples from individuals with extreme phenotypes (e.g., high and low growth rates) and identifying genetic markers associated with the trait of interest. This method provides significant cost savings compared to individual genotyping.
The combination of BSA with next-generation sequencing (BSA-seq) has revolutionized the identification of trait-related markers. By analyzing sequencing data from pooled DNA samples, researchers can identify genetic variants linked to specific traits.
Applications of BSA-seq in Aquaculture
BSA-seq has been successfully applied to several aquaculture species, including L. vannamei. In this study, researchers used BSA-seq to identify SNPs and genes associated with growth traits in L. vannamei. By analyzing two independent families, they were able to minimize the risk of false positives and identify genetic markers with high confidence.
Benefits of BSA-seq for Aquaculture Breeding
The results of the study, published in the journal Aquaculture, offer several benefits for aquaculture breeding:
- Understanding the genetic basis of growth traits: The identified SNPs and genes provide valuable insights into the genetic mechanisms underlying shrimp growth.
- Marker-assisted selection (MAS): The identified markers can be used in MAS programs to select individuals with desirable growth traits.
- Genomic selection (GS): The identified markers can also be used in GS programs to predict the breeding values of individuals based on their genomic information.
In the study, BSA-seq was employed to identify SNPs and growth-related genes in Pacific white shrimp Litopenaeus vannamei. Two independent families were used to reduce the likelihood of false positives. DNA samples from individuals with the highest and lowest body weights were pooled and sequenced.
Stay Always Informed
Join our communities to instantly receive the most important news, reports, and analysis from the aquaculture industry.
Data Analysis and Results
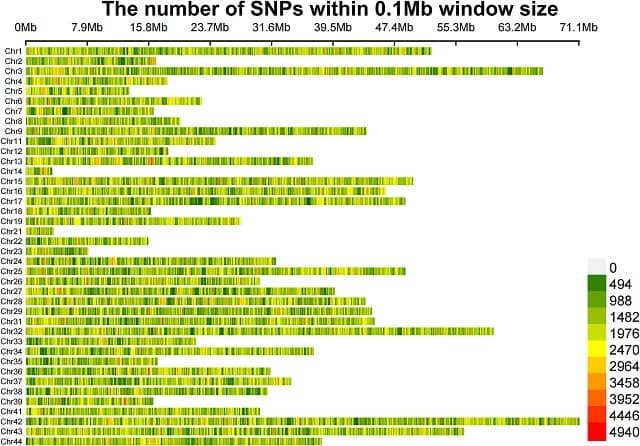
Two statistical methods, the Euclidean Distance (ED) method and the chi-square test, were used to calculate allele differences between individuals with high and low growth rates. A total of 24,325,437 SNPs were identified, and 1,299 SNPs and 163 genes were found to be associated with growth traits.
Enrichment Analysis and Candidate Genes
KEGG enrichment analysis revealed that the identified genes were significantly enriched in the phosphatidylinositol signaling and phospholipase D signaling pathways. Several genes involved in these pathways, including diacylglycerol kinase genes, phosphatidylinositol 3-kinase genes, inositol polyphosphate 5-phosphatase genes, mitogen-activated protein kinase genes, myotubularin-related protein genes, and epidermal growth factor receptor genes, were identified as potential candidate growth genes.
Implications for the Shrimp Industry
The results of this study provide valuable insights into the genetic basis of growth traits in Pacific white shrimp. The identified SNPs can be used for MAS and GS to accelerate genetic improvement in shrimp breeding programs. Additionally, the study demonstrates the efficiency and cost-effectiveness of BSA-seq for SNP identification in aquaculture species.
Conclusion
BSA-seq represents a promising approach for identifying growth-related SNPs in aquaculture species. By leveraging this technique, researchers can gain a deeper understanding of the genetic architecture of important traits and develop more efficient breeding strategies. The findings of this study contribute to ongoing efforts to improve the sustainability and profitability of shrimp aquaculture.
The study was funded by the Strategic Priority Research Program of the Chinese Academy of Sciences, the National Key Research and Development Program of China, and the Key Research and Development Program of Shandong.
Contact
Fuhua Li
Key Laboratory of Breeding Biotechnology and Sustainable Aquaculture(CAS), Institute of Oceanology, Chinese Academy of Sciences
Qingdao 266071, China.
Email: fhli@qdio.ac.cn
Reference (open access)
Bao, Z., Yu, Y., Lin, P., & Li, F. (2025). Identification of growth-related genes based on BSA in Pacific white shrimp Litopenaeus vannamei. Aquaculture, 596, 741708. https://doi.org/10.1016/j.aquaculture.2024.741708
Editor at the digital magazine AquaHoy. He holds a degree in Aquaculture Biology from the National University of Santa (UNS) and a Master’s degree in Science and Innovation Management from the Polytechnic University of Valencia, with postgraduate diplomas in Business Innovation and Innovation Management. He possesses extensive experience in the aquaculture and fisheries sector, having led the Fisheries Innovation Unit of the National Program for Innovation in Fisheries and Aquaculture (PNIPA). He has served as a senior consultant in technology watch, an innovation project formulator and advisor, and a lecturer at UNS. He is a member of the Peruvian College of Biologists and was recognized by the World Aquaculture Society (WAS) in 2016 for his contribution to aquaculture.