Advancements in genomic technologies have led to the discovery and application of DNA markers for genetic improvement of various aquaculture species.
The identification of specific genomic regions associated with important economic traits, using, for example, Genome Wide Association Studies (GWAS), allowed the discovery and incorporation of markers linked to Quantitative Trait Loci (QTL) in aquaculture breeding programs through Marker-Assisted Selection (MAS).
Researchers from the University of Chile, the University of Edinburgh, the Swedish University of Agricultural Sciences, AquaGen, and Xiamen University have published a scientific review that provides a global perspective on the development and application of genomic technologies to discover the genetic basis of complex traits and accelerate genetic progress in aquaculture species, such as salmon, trout, carp, tilapia, catfish, shrimp, scallops, and oysters.
The document also provides future perspectives on the deployment of new molecular technologies for selective breeding research in the coming years.
Genomic Resources
Genomic resources for most species that support the global aquaculture industry include high-quality reference genome assemblies and SNP panels for various species.
The study cites the status of genome sequences for important aquaculture species such as Atlantic salmon, rainbow trout, Coho salmon, carp, goldfish, Nile tilapia, catfish, pangasius, oyster, and white shrimp.
Reference genomes are critical resources for designing and generating genome-wide SNP panels, which will have a significant impact on unraveling the genetic architecture of complex traits and accelerating response to selection in aquaculture species to improve industry performance.
Genetic Architecture of Desired Traits
The goal of Genome-Wide Association Studies (GWAS) is to identify genomic regions involved in determining phenotypic variation for a particular trait.
In the aquaculture industry, traits of interest vary within species and even within the same species, being defined based on the objectives of selective breeding. However, several traits of interest exist for the selection of different species: growth, disease resistance, sexual maturation, meat quality, and tolerance to different environmental factors.
The document reports on GWAS studies in salmonids, catfish, carp, tilapia, seabass, bivalves, and shrimp.
Genomic Selection
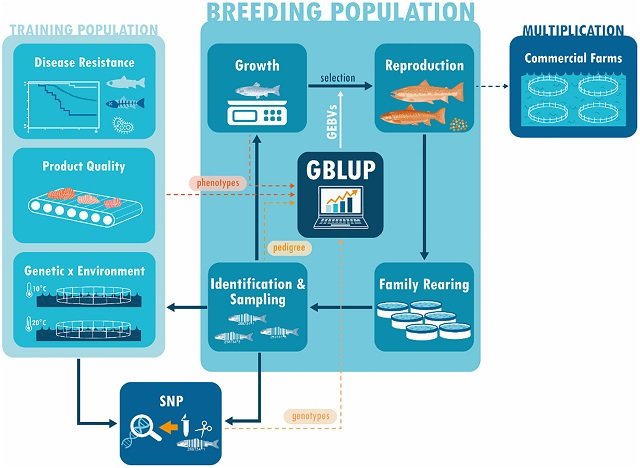
Genomic selection uses genotype markers and phenotypic data to predict genomic breeding values and has been shown to improve selection accuracy.
The main benefits of applying genomic selection in the aquaculture industry are related to the evaluation of the difficulty of measuring traits, the higher accuracy in estimating additive genetic variance, and the reduction in the generational interval.
The genomic selection strategy may be relevant for traits that are difficult or impossible to measure directly in candidate selection, such as disease resistance, carcass traits, traits limited by sex, and when genotype-environment interaction is present.
Domestication and Selection Signatures
Domestication and selective breeding have resulted in significant phenotypic changes in aquaculture species.
According to the researchers, intense selective breeding and adaptation to local environments have given rise to different lines of Atlantic salmon, Nile tilapia, rainbow trout, among other species.
The characterization of genomic regions affected by selection can allow inferences about genomic regions, functionality, and genes underlying the expression of specific traits; therefore, selection signature studies have been conducted in various species, both in wild and domestic populations.
Genome Resequencing
The availability of genome sequences for many individuals can be useful in the search for rare genetic variants associated with economic traits for the aquaculture industry.
Advancements in sequencing technologies have led to a decrease in sequencing costs, which has allowed resequencing of genomes in different aquaculture species to identify genetic variations associated with desired traits.
Conclusion
“In just two decades, the application of genomics in aquaculture breeding has gone from ‘none’ to ‘common’. Nowadays, many aquaculture improvement programs use genomic or marker-assisted selection in some way to propagate their populations or to produce eggs,” conclude the researchers.
The study was funded by the Alliance of International Science Organizations – ANSO, the National Fund for Scientific and Technological Development, and the National Natural Science Foundation of China.”
Contact
José M. Yáñez
Facultad de Ciencias Veterinarias y Pecuarias
Universidad de Chile, Santiago, Chile.
Email: jmayanez@uchile.cl
Reference (open access)
Yáñez, JM, Barría, A, López, ME, et al. Genome-wide association and genomic selection in aquaculture. Rev Aquac. 2023; 15( 2): 645- 675. doi:10.1111/raq.12750

