Biofloc Technology (BFT) has revolutionized shrimp aquaculture by fostering a healthy environment for shrimp through the power of small microbial communities. These communities, known as bioflocs, play a crucial role in maintaining water quality, improving shrimp health, and shaping the gut microbiome of these valuable crustaceans.
Despite its global adoption, a full understanding of the intricate web of microbes within bioflocs has remained elusive. This is mainly due to the great diversity of these microbes, many of which defy traditional lab-based cultivation methods.
The Microbial Universe
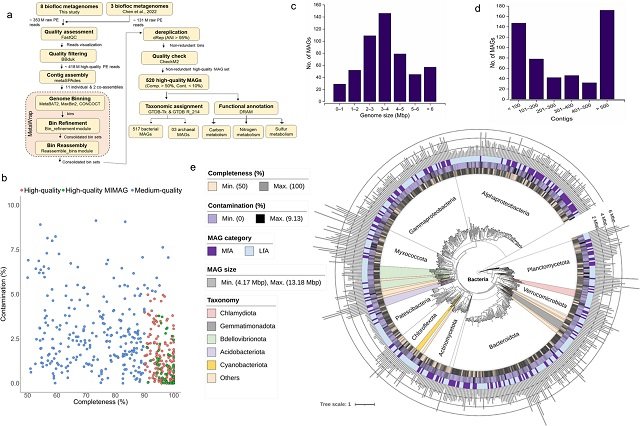
A recent groundbreaking study published in the journal mSystems by scientists from Inha University sheds light on this hidden world. The researchers employed a cutting-edge technique called ‘genome-centric metagenomics’ to delve into the biofloc-associated microbiota (FAB community) in shrimp aquaculture systems in South Korea and China. This approach allowed them to reconstruct an astonishing 520 metagenome-assembled genomes (MAGs), essentially blueprints of individual microbes, encompassing both bacteria and archaea.
A Treasure Trove of Diversity
The analysis revealed a fascinating insight into the FAB community. Two prominent bacterial groups, Pseudomonadota and Bacteroidota, emerged as key members. Surprisingly, nearly 93% of the recovered MAGs defied species-level classification. This points to a vast reservoir of uncharacterized microbial diversity within the FAB community, awaiting exploration.
Unveiling Functional Power
By analyzing the functional potential encoded within these MAGs, the researchers unraveled the incredible metabolic capabilities of the FAB community. These microbes possess a diverse repertoire for degrading complex carbohydrates, significantly contributing to the carbon cycle within the aquaculture system.
Masters of Biogeochemistry
Perhaps the most exciting finding of the study lies in the versatility of the FAB community in nitrogen and sulfur metabolisms. The study identified genes associated with crucial processes like ammonium assimilation, nitrification, denitrification, and thiosulfate and sulfur oxidation. This translates into the FAB community’s ability to thrive in both aerobic (oxygen-rich) and anaerobic (oxygen-poor) environments, ensuring the efficient removal of toxic waste products such as ammonia and sulfide. Notably, the research indicates the absence of genes linked to less desirable pathways like heterotrophic nitrification and sulfate reduction.
Rhodobacteraceae: The Stars
Among this diverse microbial tapestry, members of the Rhodobacteraceae family stood out as the most abundant and metabolically versatile. Their presence suggests a vital role in driving key biogeochemical processes within the biofloc system.
Impact on Sustainable Aquaculture
This groundbreaking study not only expands the current collection of microbial genomes from aquaculture environments but also provides valuable insights into the functional capabilities of the FAB community. By illuminating the intricate web of interactions between various microbial taxa and their contributions to biogeochemical cycles, this research paves the way for future advancements in biofloc technology.
Moreover, the knowledge gained from this study paves the way for:
Stay Always Informed
Join our communities to instantly receive the most important news, reports, and analysis from the aquaculture industry.
- The development of specific probiotics to enhance the beneficial biofloc community.
- Optimization of BFT systems to improve water quality and shrimp health.
- Identification of new microbial strains with potential applications in aquaculture.
Conclusion
This study delves into the intricate microbial community found in biofloc aquaculture systems, analyzing its composition and metabolic functions. Here are some key findings:
Microbial Diversity and Composition
- Vast uncultivated diversity: The study identified a wide range of microbial species within the biofloc-associated microbiota (FAB community). However, more than 90% of these species remain uncultivated, highlighting the importance of metagenomic approaches to understand this unique ecosystem.
- Close relationship with shrimp gut microbiota: The FAB community shares a great resemblance to the shrimp gut microbiota. This suggests a potential impact on shrimp health and immunity, likely due to the shrimp’s reliance on bioflocs as a food source.
- Key members of the FAB community: The study identified key microbial taxa consistently present in the FAB community. These include members of the families Rhodobacteraceae, Halieaceae, Flavobacteriaceae, Saprospiraceae, Cyclobacteriaceae, Planctomycetaceae, Microbacteriaceae, and Nannocystaceae.
Metabolic Functions of the FAB Community
- Carbon degradation: The FAB community excels at degrading complex organic carbon, with members of the phylum Bacteroidota, particularly Flavobacteriaceae and Saprospiraceae, playing a key role.
- Nitrogen metabolism: While the FAB community primarily relies on heterotrophic ammonium assimilation for nitrogen removal, it also exhibits potential for denitrification and dissimilatory nitrate reduction to ammonium (DNRA).
- Sulfur oxidation: A significant portion of the FAB community participates in sulfur oxidation, contributing to water quality maintenance. Rhodobacteraceae and Arenicellales are particularly active in this process.
Overall, this study provides valuable insights into the microbial diversity and metabolic capabilities of the FAB community in biofloc aquaculture systems. The findings have significant implications for enhancing the sustainability and efficiency of aquaculture.
The research was conducted under the High Seas Bioresources Program of the Korea Institute of Marine Science & Technology Promotion (KIMST), funded by the Ministry of Oceans and Fisheries, the Mid-Career Research Program, and the Science Research Center Program through the National Research Foundation (NRF), funded by the Ministry of Science and ICT.
Contact
Jang-Cheon Cho
Department of Biological Sciences and Bioengineering
Center for Molecular and Cell Biology, Inha University
Incheon, South Korea
Email: chojc@inha.ac.kr
Reference (open access)
Rajeev M, Jung I, Kang I, Cho J. 2024. Genome-centric metagenomics provides insights into the core microbial community and functional profiles of biofloc aquaculture. mSystems0:e00782-24.https://doi.org/10.1128/msystems.00782-24
Editor at the digital magazine AquaHoy. He holds a degree in Aquaculture Biology from the National University of Santa (UNS) and a Master’s degree in Science and Innovation Management from the Polytechnic University of Valencia, with postgraduate diplomas in Business Innovation and Innovation Management. He possesses extensive experience in the aquaculture and fisheries sector, having led the Fisheries Innovation Unit of the National Program for Innovation in Fisheries and Aquaculture (PNIPA). He has served as a senior consultant in technology watch, an innovation project formulator and advisor, and a lecturer at UNS. He is a member of the Peruvian College of Biologists and was recognized by the World Aquaculture Society (WAS) in 2016 for his contribution to aquaculture.