Bivalves, such as oysters, scallops, and clams, are an economic cornerstone of global aquaculture, accounting for over half of the mariculture production in industry powerhouses like China. For producers, enhancing growth rates is a primary objective, as it directly influences harvest yield and commercial value. However, genetic progress has been constrained by a critical bottleneck: the measurement of traits like size and weight.
Traditionally, this process is performed manually using calipers and scales—a slow, laborious, and error-prone method that undermines the precision of broodstock selection and hinders genetic advancement.
Now, a scientific study in the journal Aquaculture Reports introduces an innovative solution: an automated phenotyping system that integrates multiple cutting-edge technologies. This system promises not only to dramatically accelerate the process but also to boost accuracy to unprecedented levels, heralding a new era for selective bivalve breeding.
An integrated system for precision aquaculture
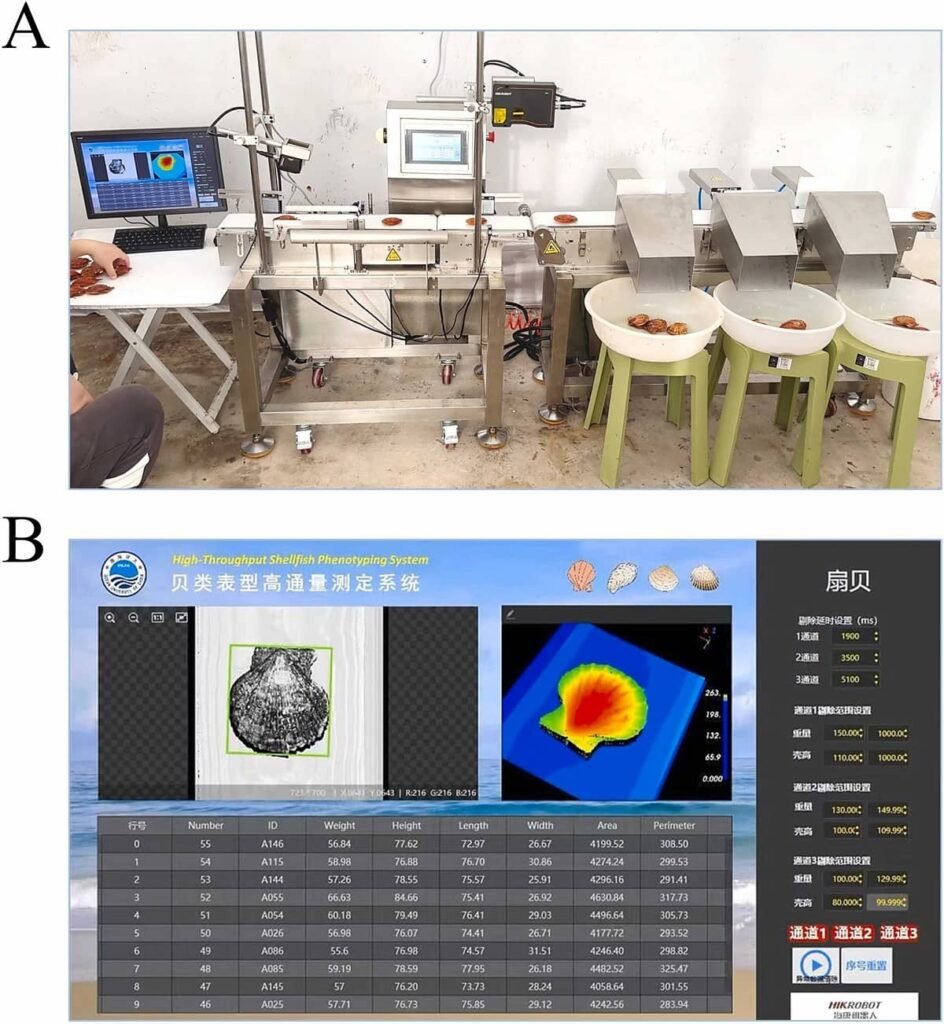
Researchers from the Ocean University of China and the Qingdao Marine Science and Technology Center have developed a high-throughput phenotyping system that combines four key modules into a streamlined assembly line: 3D laser imaging, dynamic weighing, automated sorting, and barcode identification. This integrated platform is engineered to rapidly and accurately measure growth traits, classify individuals, and log their data within a seamless workflow.
High-precision size measurement via 3D laser imaging
At the core of the system is a 3D laser imaging platform that scans bivalves as they travel on a conveyor belt. A laser sensor projects a line onto the shell, and through triangulation analysis, the software calculates key dimensions in real-time, including shell height, length, width, area, and circumference. The results demonstrate exceptional precision, with the platform’s measurement error ranging between 0.01 and 0.30 mm. A comparison with manual caliper measurements across five different species revealed a near-perfect correlation (R2>0.91 in all cases), and the entire process for an individual takes just 1.2 seconds.
Dynamic weighing: Speed and accuracy in motion
To overcome the slow pace of static scales, the system incorporates a dynamic weighing platform that measures the weight of bivalves while they are in motion. Even with individuals moving at speeds up to 1.0 m/s, the platform achieved over 99% accuracy compared to a static electronic scale.
Automated sorting for broodstock selection
After being measured and weighed, the bivalves move to an automated sorting platform. Using real-time data, the system activates pushers that guide each individual to the appropriate discharge gate based on criteria preset by the operator (e.g., Grade 1: >40g; Grade 2: 20-40g; Grade 3: <20g). Tests on scallops and oysters showed a 100% sorting accuracy, ensuring every animal was assigned to the correct category.
Individual identification: Precise tracking with QR codes
To link phenotypic data to each bivalve, a robust and efficient tagging method was developed using small PVC tags with unique QR codes, which are affixed to the shell with a seawater-resistant superglue. The durability of this method is remarkable; after six months in sea cages, 99.67% of the tagged bivalves retained at least one tag, and the QR codes remained legible to a high-resolution scanner. This enables reliable growth tracking over time and seamless integration of phenotypic and genotypic data.
Striking results: 11 times faster than manual methods
The integrated system’s most impressive advantage is its efficiency. While manually measuring, weighing, sorting, and identifying a single bivalve takes an average of 18.2 seconds, the automated system completes all tasks in just 1.6 seconds per individual. This makes the system approximately 11 times faster than manual operations, allowing it to process over two thousand bivalve samples per hour and meet the demands of large-scale breeding programs.
Stay Always Informed
Join our communities to instantly receive the most important news, reports, and analysis from the aquaculture industry.
Implications for genetic improvement and the aquaculture industry
The implementation of this automated phenotyping system has the potential to transform selective breeding programs for bivalves. By providing large-scale, high-precision phenotypic data rapidly, the technology enables:
- Accelerated genetic progress: The selection of superior broodstock becomes more precise and efficient, which can significantly improve growth traits in future generations.
- Enhanced genomic selection: The wealth of accurate phenotypic data enriches genome-wide association studies (GWAS), simplifying the identification of growth-related genes.
- Versatile application: The system has proven effective for bivalves with diverse morphologies, from regularly shaped scallops to irregularly shaped oysters, suggesting its broad applicability across commercially important species.
Conclusion
The development of this automated phenotyping system marks a significant breakthrough for bivalve aquaculture. By overcoming the limitations of manual methods, this integrated platform provides a fast, accurate, and efficient tool for measuring, sorting, and identifying bivalves on an industrial scale. Its ability to accelerate breeding programs and improve data management positions it as a key technology that will contribute to a more productive and sustainable aquaculture industry.
Contact
Xiaoli Hu
Ministry of Education Key Laboratory of Marine Genetics and Breeding, College of Marine Life Science, Ocean University of China
Qingdao 266003, China
Email: hxl707@ouc.edu.cn
Reference (open access)
Kong, X., Huang, S., Wang, X., Liang, H., Hu, C., Wang, Y., Bao, Z., & Hu, X. (2025). Development of automated phenotyping system for growth traits in bivalves. Aquaculture Reports, 44, 103019. https://doi.org/10.1016/j.aqrep.2025.103019
Editor at the digital magazine AquaHoy. He holds a degree in Aquaculture Biology from the National University of Santa (UNS) and a Master’s degree in Science and Innovation Management from the Polytechnic University of Valencia, with postgraduate diplomas in Business Innovation and Innovation Management. He possesses extensive experience in the aquaculture and fisheries sector, having led the Fisheries Innovation Unit of the National Program for Innovation in Fisheries and Aquaculture (PNIPA). He has served as a senior consultant in technology watch, an innovation project formulator and advisor, and a lecturer at UNS. He is a member of the Peruvian College of Biologists and was recognized by the World Aquaculture Society (WAS) in 2016 for his contribution to aquaculture.