Success in large-scale microalgae production, such as Desmodesmus armatus, depends on more than just light, nutrients, or reactor design. There is an invisible, microscopic universe —a complex ecosystem known as the microbiome —that lives alongside the algae and plays a decisive role in the health and productivity of the culture. A recent study, published in Bioresource Technology by scientists from the Institute of Marine Sciences (ICM) the University of Murcia and the University of Almería, has delved into this intricate network of interactions to reveal which microorganisms act as allies and which as enemies.
The findings offer a valuable roadmap for optimizing industrial production processes, especially in promising areas such as wastewater treatment and sustainable biomass generation.
The microbial universe in raceway reactors
Microalgae are a cornerstone of the circular bioeconomy, capable of converting sunlight and CO2 into high-value bioplastics, biofuels, and nutritional supplements. However, bringing their production to an industrial scale faces a critical challenge: culture stability. Drops in productivity or the total collapse of a culture are often due to imbalances in its microbiome, where harmful microorganisms gain ground over beneficial ones.
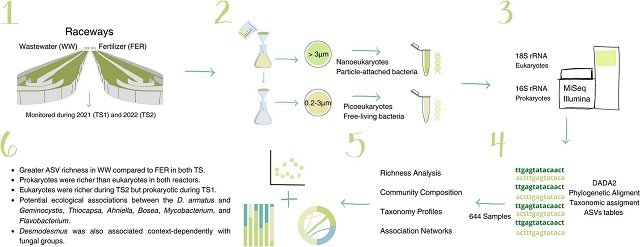
To understand these dynamics, a research team monitored the microbial communities in two large-scale raceway reactors over two 8-month periods (in 2021 and 2022), both inoculated with the microalga Desmodesmus armatus. The main difference between them was their “diet”: one was fed with urban wastewater (WW), while the other used clean water supplemented with fertilizers (FER).
Using the 16S and 18S rRNA metabarcoding technique, the scientists analyzed the diversity and composition of the prokaryotic (bacteria) and eukaryotic (such as fungi and other protists) communities growing alongside the microalga.
Who lives alongside Desmodesmus armatus?
The study revealed an astonishing and complex microbial diversity. Some of the most prominent findings on the composition of these communities were:
- Higher richness in wastewater: The reactor using wastewater (WW) showed a significantly higher species richness (number of genetic variants or ASVs) than the reactor with fertilizers (FER), for both bacteria and eukaryotes. This suggests that the composition of the input water is a key factor in shaping the microbiome.
- Bacterial dominance: In both reactors, the richness of bacteria was consistently higher than that of eukaryotes. The dominant bacterial groups included Alphaproteobacteria, Gammaproteobacteria, Actinobacteria, and Bacteroidia.
- Fungal communities: Fungi represented a significant part of the eukaryotic richness, making up approximately one-third of the ASVs in this category. Among them, groups known to be microalgae parasites were identified, such as Rozellomycota and Aphelidiomycota.
Identifying the “Good Guys”: Bacteria that promote a healthy culture
The analysis made it possible to correlate the presence of certain microorganisms with “healthy” culture conditions (defined as an abundance of D. armatus greater than 70%). In this group of potential “allies,” several bacteria stood out:
- Geminocystis (ASV_2): This cyanobacterium was consistently associated with healthy cultures in both types of reactors and over the two years of the study. The recurrent presence of the cyanobacterium in optimal conditions and its positive association in network analyses point to it as a potential benefactor for D. armatus.
- Thiocapsa: This genus of Gammaproteobacteria was linked to healthy conditions in the wastewater reactor. As a phototrophic bacterium, it could benefit Desmodesmus by participating in carbon, nitrogen, and phosphorus recycling.
- Ahniella (ASV_63): Also a Gammaproteobacteria, it showed a positive interaction with the microalga and appeared in healthy conditions, especially in wastewater treatment, where it is a common bacterium in microalgal biofilms.
- Bosea: Belonging to the class Rhizobiales, it was found in healthy conditions in the fertilizer reactor and, to a lesser extent, in the wastewater reactor. Bacteria from this group are common in the phycosphere (the area surrounding the algae) of industrial cultures.
Detecting the “Bad Guys”: Bacterial and fungal pathogens
Similarly, the study identified microorganisms associated with “unhealthy” conditions (with an abundance of D. armatus below 20%), revealing the main suspects for harming the culture.
- Mycobacterium: Various variants (ASVs) of this bacterium consistently appeared in unhealthy cultures in both reactors and over time. The negative association of Mycobacterium with D. armatus in network analyses underscores its potentially detrimental effect, a novel finding as its impact on microalgae cultures had not been previously documented.
- Flavobacterium: Although its role can be complex, certain variants of this bacterium were associated with unhealthy conditions and showed negative interactions. Some species of Flavobacterium are known to attack microalgae, either through direct contact or by releasing compounds that degrade their cell walls.
- Parasitic fungi: The study reaffirmed the harmful role of several fungal groups and, for the first time, identified others as possible threats to D. armatus:
- Paraphelidium tribonemae (ASV 8) and Aphelidium parallelum (ASV_39): They were identified for the first time as potential parasites of D. armatus, being linked to unhealthy conditions.
- Aphelidium desmodesmi (ASV_6) and Rhizophydium sp. (ASV 37): They confirmed their known role as pathogens, appearing in weakened cultures.
- Rozellomycota: Different variants of this group, known microalgae parasites, were negatively associated with the health of the culture in most cases.
It is interesting to note that the role of some fungi appears to be context-dependent. For example, one variant of Aphelidium desmodesmi (ASV_104) was associated with unhealthy conditions in three of the four scenarios, but with healthy conditions in one, demonstrating the complexity of these interactions.
Stay Always Informed
Join our communities to instantly receive the most important news, reports, and analysis from the aquaculture industry.
Implications for the future of microalgae production
This study not only maps the complex microbial community in microalgae reactors but also provides crucial information for the management and optimization of these systems. The ability to early identify an increase in the population of pathogens like Mycobacterium or parasitic fungi could serve as an early-warning signal to prevent a culture collapse.
Likewise, understanding which beneficial bacteria like Geminocystis thrive alongside D. armatus opens the door to designing “probiotic” strategies for microalgae, where specific microbial consortia could be inoculated to improve the resilience and productivity of the system.
In summary, the path toward more robust and efficient microalgae production inevitably involves understanding and managing the tiny but powerful microbiome that accompanies them.
The study was funded by the European Union’s Horizon 2020 Research and Innovation Programme through the PRODIGIO project: “Developing early-warning systems for improved microalgae PROduction and anaerobic DIGestION”, and by the INCEPTION project “In-depth characterization of microalgal crop genomics for sustainable biomass production and circular bioeconomy” funded by the Spanish Research Agency (AEI).
Contact
Judith Traver-Azuara
Institute of Marine Sciences (ICM), CSIC
Barcelona 08003, Spain
Email: judithtraverazuara@icm.csic.es
Reference (open access)
Traver-Azuara, J., Giner, C. R., García-Comas, C., Sánchez-Zurano, A., Ciardi, M., Acién, G., Bondarenko, S., Obiol, A., Massana, R., Sala, M. M., Logares, R., & Cermeño, P. (2025). Complex interplay between the microalgae and their microbiome in production raceways. Bioresource Technology, 432, 132650. https://doi.org/10.1016/j.biortech.2025.132650
Editor at the digital magazine AquaHoy. He holds a degree in Aquaculture Biology from the National University of Santa (UNS) and a Master’s degree in Science and Innovation Management from the Polytechnic University of Valencia, with postgraduate diplomas in Business Innovation and Innovation Management. He possesses extensive experience in the aquaculture and fisheries sector, having led the Fisheries Innovation Unit of the National Program for Innovation in Fisheries and Aquaculture (PNIPA). He has served as a senior consultant in technology watch, an innovation project formulator and advisor, and a lecturer at UNS. He is a member of the Peruvian College of Biologists and was recognized by the World Aquaculture Society (WAS) in 2016 for his contribution to aquaculture.