The global fishing and aquaculture industry is expanding rapidly. With total production reaching 223.2 million tons in 2022 and trade exceeding $160 billion annually, the demand for high-quality protein has never been higher. To meet this demand sustainably, modern aquaculture must optimize feed efficiency, enhance reproduction, regulate growth, and decipher disease resistance mechanisms.
The key to achieving these advancements lies in data. Specifically, in “multi-omic” data: the combined information from an organism’s genomics, transcriptomics, epigenetics, and proteomics. Until now, however, this vital data for fish species was fragmented, scattered across various publications or incomplete databases.
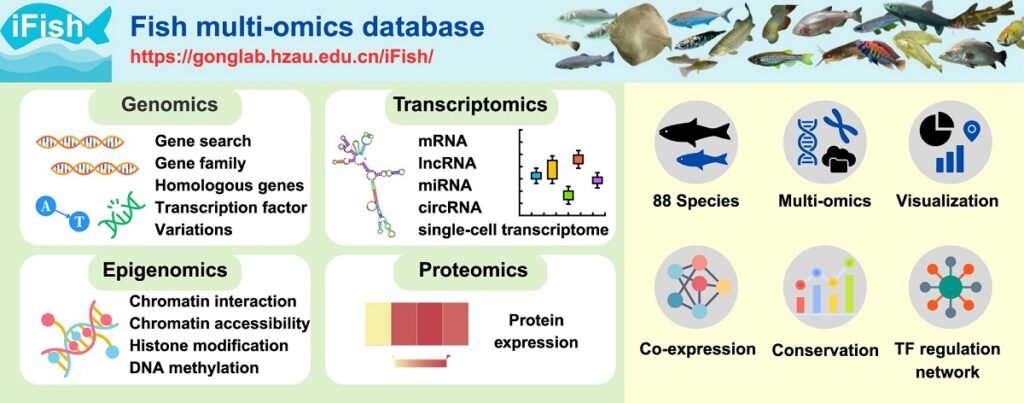
To solve this problem, a team of researchers from Huazhong Agricultural University has developed iFish, a comprehensive platform designed to be the most complete multi-omic resource for piscine research to date.
- 1 Key conclusions
- 2 The challenge of omics data in aquaculture
- 3 iFish: An integral solution for research
- 4 What does the iFish database contain?
- 5 The power of non-coding RNA (ncRNA): A key advantage of iFish
- 6 More than just data: iFish tools and functionalities
- 7 A case study: Exploring sex determination in zebrafish
- 8 Implications for the future of aquaculture
- 9 Conclusion: The future is multi-omic
- 10 Entradas relacionadas:
Key conclusions
- iFish has been developed as the most comprehensive platform to date, integrating multi-omic data (genome, transcriptome, epigenome, and proteome) from 88 fish species.
- The database centralizes a massive amount of information, including 137 million single nucleotide polymorphisms (SNPs), 2.7 million gene annotations, and 192,107 transcription factors.
- A distinctive strength of iFish is its rich repository of non-coding RNAs (ncRNAs), such as lncRNAs and miRNAs, whose function is crucial for fish biology but was previously under-characterized.
- The platform offers user-friendly tools for visualization, download, and functional analysis, including gene co-expression and transcription factor regulatory networks.
- iFish is designed to accelerate genetic research and genetic improvement in aquaculture, facilitating the discovery of genes linked to growth, disease resistance, and sex determination.
The challenge of omics data in aquaculture
Fish are the most diverse group of vertebrates and play a fundamental role in global nutrition and ecological balance. Investigating their biology to improve aquaculture requires a deep understanding of their genomic architecture, gene regulatory networks, and epigenetic mechanisms.
In recent years, scientists have used different omics approaches to study, for example, genetic markers (SNPs) related to growth in yellow catfish or how DNA methylation influences X and Y sperm.
However, existing databases were limited. Some focused only on zebrafish, others covered few species, or they specialized in only one type of data, such as microRNAs (miRNAs) or transposable elements. A platform that integrated everything was missing.
According to the study’s authors, previous databases were “not sufficiently comprehensive” in terms of species and data types. Crucial aspects such as genetic variation, non-coding RNA (ncRNA) identification, and epigenomic datasets were insufficient for most species.
iFish: An integral solution for research
To fill this gap, iFish was created (accessible at https://gonglab.hzau.edu.cn/iFish/), a rigorously curated database that integrates multiple layers of omic information for 88 fish species.
This massive data collection, exceeding 40 TB, includes both model species (like zebrafish) and species of high economic importance, including:
Stay Always Informed
Join our communities to instantly receive the most important news, reports, and analysis from the aquaculture industry.
- Atlantic salmon (Salmo salar)
- Atlantic cod (Gadus morhua)
- Rainbow trout (Oncorhynchus mykiss)
- Gilt-head bream (Sparus aurata)
- Grass carp (Ctenopharyngodon idella)
- Largemouth bass (Micropterus salmoides)
- Yellow catfish (Pelteobagrus fulvidraco)
iFish is not just a repository; it is an analysis platform that allows users to interactively browse, visualize, and download data.
What does the iFish database contain?
The iFish platform is organized into modules covering four main areas of molecular biology, processing a total of 13,377 sequencing samples:
Genomics (The genetic ‘map’)
This module provides the DNA foundation. It integrates 88 genomic assemblies and data from 884 whole-genome sequencing (WGS) projects. From this, the researchers identified:
- 137.04 million SNPs (single nucleotide polymorphisms).
- 45.52 million InDels (insertions/deletions).
- 2.7 million gene annotation entries.
- 192,107 transcription factors (TFs), which are the proteins that “turn on” or “turn off” genes.
- Homologous gene analysis across species.
Transcriptomics (Which genes are being ‘used’)
This section analyzes gene expression (RNA). iFish processed 9,797 bulk RNA-seq, 293 single-cell RNA-seq (scRNA-seq), and 840 microRNA-seq datasets. This allowed for the quantification of:
- 1.87 million mRNAs (messenger RNAs).
- 287,873 lncRNAs (long non-coding RNAs).
- 197,554 circRNAs (circular RNAs).
- 6,068 mature miRNAs (microRNAs).
Epigenomics (How the ‘map’ is regulated)
Epigenetics studies the chemical switches that, without changing the DNA, control which genes are activated. iFish integrates 1,563 epigenomic datasets to map:
- Histone modifications.
- Chromatin accessibility (which parts of the DNA are “open” for use).
- DNA methylation.
- 3D chromatin interactions (how DNA is organized in space).
Proteomics (The functional ‘machines’)
Finally, proteomics analyzes the proteins, the final products of genes. This module includes 50 projects for profiling protein expression.
The power of non-coding RNA (ncRNA): A key advantage of iFish
One of the strengths that distinguishes iFish is its rich repository of information on non-coding RNAs (ncRNAs).
In recent years, scientists have discovered that ncRNAs, once considered “junk DNA,” are actually crucial biological regulators. In fish, ncRNAs have been shown to play vital roles in development and immune response. For example, the study mentions how a specific lncRNA (AANCR) promotes antiviral responses, and how a miR-722 helps modulate the immune response against bacteria in flounder.
Despite their importance, the expression profiles and functions of ncRNAs in fish were poorly characterized, and a comprehensive platform was lacking. iFish addresses this systematically, identifying and characterizing the expression landscape of lncRNAs, circRNAs, and miRNAs, and even investigating their conservation across different fish species.
More than just data: iFish tools and functionalities
The true power of iFish lies not just in the quantity of data, but in how it allows researchers to use it. The platform offers functional analysis tools that convert raw numbers into biological knowledge.
Two of the most powerful tools are:
- Gene Co-expression Networks: iFish has calculated 289 million pairs of genes that “turn on” and “turn off” together across 19 species. This allows a researcher to search for a gene of interest and instantly discover which other genes it is collaborating with.
- TF Regulatory Networks: The database maps 281 million regulatory pairs between transcription factors (TFs) and their target genes in 46 species. This helps decipher how genetic networks are controlled.
A case study: Exploring sex determination in zebrafish
To demonstrate the utility of iFish, the researchers conducted a case study on the dmrt1 gene in zebrafish.
Sex determination is crucial in aquaculture, as culturing single-sex (monosex) populations can be highly valuable. The dmrt1 gene is known to be a key player in male sexual development in many fish.
Using iFish, the researchers:
- Searched for the “dmrt1” gene in the gene search module.
- The platform displayed the gene’s information, its location on the genome (visualized with JBrowse), and its expression profile across different tissues.
- As expected, the results showed high expression of dmrt1 specifically in testicular tissue, confirming previous reports and validating the database’s accuracy.
- Furthermore, they could use the “Function” tool to instantly generate a co-expression network, showing all genes that work together with dmrt1 in the zebrafish testis.
Implications for the future of aquaculture
iFish represents a significant leap forward for piscine research. By unifying genomic, transcriptomic, epigenetic, and proteomic data under a single, accessible platform, it drastically reduces the time and effort required to perform complex analyses.
For aquaculture professionals and geneticists, this has direct practical implications:
- Accelerated Genetic Improvement: Breeders can use iFish to more quickly identify genetic markers (SNPs) and genes associated with desirable traits, such as faster growth or better feed conversion.
- Health and Disease: Researchers can explore the gene expression profiles (like ncRNAs) and epigenetic changes (like DNA methylation) that occur when a fish responds to a pathogen or environmental stress.
- Reproductive Optimization: The ability to investigate sex determination networks (like the dmrt1 example) facilitates the development of monosex populations.
- Evolutionary Research: The platform allows for conservation analysis across species, helping to understand the evolution of fish.
Conclusion: The future is multi-omic
The iFish database is, to date, the most comprehensive platform that systematically integrates, analyzes, and stores multi-omic data for fish. By providing a user-friendly “one-stop shop,” the creators hope iFish will serve as a fundamental resource for advancing genetic, transcriptional regulatory, and epigenetic research.
For an aquaculture industry that depends on scientific innovation to meet growing global protein demands, tools like iFish are not just an academic advancement; they are essential drivers for genetic improvement and the practical applications that will define the sustainable future of the sector.
Contact
Ze-Xia Gao
1 Hubei Hongshan Laboratory, College of Informatics, Huazhong Agricultural University
Wuhan 430070, China
Email: gaozx@mail.hzau.edu.cn
Reference (open access)
Ruoyan Liu, Qin Tang, Peng Sun, Weiwei Jin, Yanbo Yang, Xiaohong Wu, Wen Cao, Xiaohui Niu, Jing Gong, Ze-Xia Gao, iFish: a comprehensive multi-omics database for fish genetics, transcriptional regulation, and epigenetic research, Nucleic Acids Research, 2025;, gkaf1026, https://doi.org/10.1093/nar/gkaf1026
Editor at the digital magazine AquaHoy. He holds a degree in Aquaculture Biology from the National University of Santa (UNS) and a Master’s degree in Science and Innovation Management from the Polytechnic University of Valencia, with postgraduate diplomas in Business Innovation and Innovation Management. He possesses extensive experience in the aquaculture and fisheries sector, having led the Fisheries Innovation Unit of the National Program for Innovation in Fisheries and Aquaculture (PNIPA). He has served as a senior consultant in technology watch, an innovation project formulator and advisor, and a lecturer at UNS. He is a member of the Peruvian College of Biologists and was recognized by the World Aquaculture Society (WAS) in 2016 for his contribution to aquaculture.