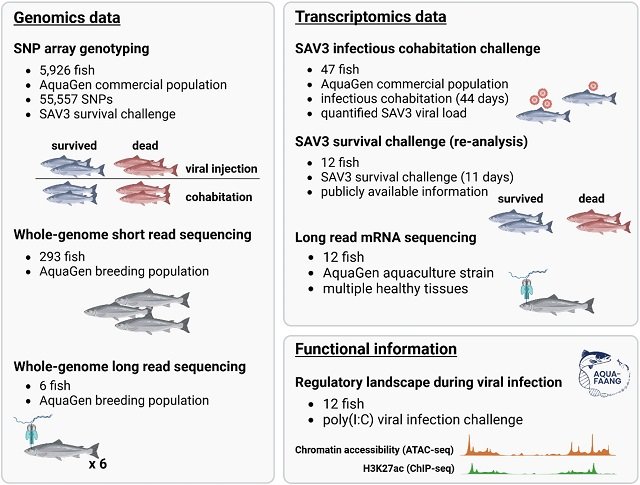
New research has shed light on the complex genetic puzzle of resistance to Pancreas Disease (PD) in Atlantic salmon. Scientists have identified a specific region in the salmon genome and three candidate genes that are decisive in the fish’s response to the salmonid alphavirus (SAV), the causative agent of this devastating disease.
These findings, published in the journal BMC Genomics by scientists from the Norwegian University of Life Sciences, AquaGen AS, and the University of Aberdeen, not only confirm the importance of genetics in the fight against PD but also open the door to developing much more precise and effective management strategies and genomic selection programs for the salmon farming industry.
- 1 A persistent challenge for salmon farming
- 2 Mapping resistance: A deep genomic dive
- 3 Identifying the genes for PD resistance
- 4 Three “gig1-like” genes: The main candidates
- 5 An unexpected paradox: More gene expression, less resistance
- 6 Implications for aquaculture: Towards more precise genomic selection
- 7 Conclusion: A more resilient future for salmon
- 8 Entradas relacionadas:
A persistent challenge for salmon farming
Pancreas Disease is one of the biggest health challenges for Atlantic salmon farming. Caused by the salmonid alphavirus (SAV), this pathology leads to severe inflammation and necrosis of the pancreas, resulting in loss of appetite, lethargy, and increased susceptibility to secondary infections.
PD outbreaks can cause mortality rates of up to 28%, and although some fish survive, they are often left with stunted growth, which negatively impacts production and animal welfare. While vaccines and selective breeding have helped mitigate the problem, their effectiveness is only partial. Therefore, thoroughly understanding the genetic basis of resistance is fundamental to developing sustainable solutions.
Mapping resistance: A deep genomic dive
To decipher the genetic code of PD resistance, researchers conducted a large-scale study using cutting-edge genomic technologies.
The study analyzed genotype and survival data from 5,628 Atlantic salmon from three independent viral challenge trials conducted between 2018 and 2020. To simulate diverse conditions, the fish were infected with the SAV3 subtype, the most aggressive in Norway, using two different methods:
- Peritoneal injection (IP): A direct and controlled infection method.
- Cohabitation (CH): A method that mimics the natural transmission of the virus on a farm, where healthy fish share water with infected individuals.
Enhancing the analysis with advanced genomics
The main breakthrough of this study was the use of genotype imputation techniques. The researchers started with a panel of approximately 55,000 genetic markers (SNPs) and, using a reference set of 293 complete genomes, were able to infer and expand the dataset to over 938,000 SNPs. This high-resolution genetic “map” allowed for a much more precise localization of the regions associated with disease resistance.
Additionally, long-read sequencing technologies were used to identify more complex genetic variants, such as insertions, deletions, and microsatellites, which standard technologies often fail to detect.
Identifying the genes for PD resistance
The analysis of this enormous amount of genomic data led to a revealing discovery about the genetic architecture of the response to PD.
Stay Always Informed
Join our communities to instantly receive the most important news, reports, and analysis from the aquaculture industry.
Two genetic regions (QTLs) in Focus, but one is the star
The study confirmed the presence of two Quantitative Trait Loci (QTLs), which are regions of the genome that affect a particular trait, associated with PD resistance:
- A major QTL on chromosome Ssa03: This region showed a very strong association with disease survival, regardless of the infection method (IP or CH).
- A secondary QTL on chromosome Ssa07: Interestingly, this region only showed a significant association in the fish infected by direct injection (IP), but not in the cohabitation group. This suggests that the Ssa07 QTL might be involved in a specific immune response activated by the artificial infection route, while the Ssa03 QTL is the main player in overall disease resistance.
Three “gig1-like” genes: The main candidates
Thanks to the high-resolution analysis, the researchers were able to narrow down the vast region of the main QTL on Ssa03 to a fragment of only 45 kilobases (kb). Within this small portion of the genome are six genes.
To determine which of these was the true culprit, gene expression data (transcriptomics) from fish with high and low viral loads were analyzed. The results were conclusive: only three copies of a gene known as gig1-like showed a significant response to SAV3 infection. This positions them as the most likely causal candidates for PD resistance.
An unexpected paradox: More gene expression, less resistance
One of the most intriguing findings of the study is the relationship between the expression of these genes and the disease. Contrary to what might be expected, the analyses showed that higher expression of the gig1-like genes correlated with a higher viral load and lower fish survival.
This suggests that the gig1-like genes, which are induced by the fish’s immune response to the virus, might function more as an indicator of viral replication than as a viral suppression mechanism. In other words, high activity of these genes seems to be a sign that the virus is winning the battle in the fish’s body.
Implications for aquaculture: Towards more precise genomic selection
These discoveries have transformative potential for genetic improvement programs in salmon aquaculture.
The identification of the three gig1-like genes and specific genetic variants within them—including single-nucleotide polymorphisms (SNPs) and microsatellites —provides geneticists with much more specific and reliable targets for selection. Instead of selecting based on a broad and poorly defined QTL region, it is now possible to focus on the genetic markers that have a direct functional impact on the disease response.
Incorporating this information into breeding programs will allow for the selection of broodstock with the genetic variants associated with greater resistance, accelerating genetic progress and contributing to the production of more robust and healthy salmon populations.
Conclusion: A more resilient future for salmon
This study represents a giant step forward in our understanding of the genetic basis of resistance to Pancreas Disease. By combining data from large-scale viral challenges with the most advanced genomic tools, researchers have moved from identifying broad genomic regions to pinpointing specific genes and the variants within them that orchestrate the response to SAV infection.
The road to salmon that are completely resistant to PD is still long, but these findings provide a detailed and precise map that will undoubtedly strengthen disease management strategies and promote a more sustainable and resilient aquaculture.
Contact
Domniki Manousi
Centre for Integrative Genetics, Department of Animal and Aquacultural Sciences, Faculty of Biosciences, Norwegian University of Life Sciences
Oluf Thesens vei 6, Ås, 1433, Norway
Email: domniki.manousi@nmbu.no
Reference (open access)
Manousi, D., Jaskula, D.M., Grammes, F. et al. Dissecting the genetic basis of response to salmonid alphavirus in Atlantic salmon. BMC Genomics 26, 657 (2025). https://doi.org/10.1186/s12864-025-11735-2
Editor at the digital magazine AquaHoy. He holds a degree in Aquaculture Biology from the National University of Santa (UNS) and a Master’s degree in Science and Innovation Management from the Polytechnic University of Valencia, with postgraduate diplomas in Business Innovation and Innovation Management. He possesses extensive experience in the aquaculture and fisheries sector, having led the Fisheries Innovation Unit of the National Program for Innovation in Fisheries and Aquaculture (PNIPA). He has served as a senior consultant in technology watch, an innovation project formulator and advisor, and a lecturer at UNS. He is a member of the Peruvian College of Biologists and was recognized by the World Aquaculture Society (WAS) in 2016 for his contribution to aquaculture.