Harmful algal blooms (HABs), also known as “red tides,” represent a growing and devastating threat to aquaculture, fisheries, and the health of marine ecosystems worldwide. The massive accumulation of phytoplankton can produce neurotoxins, create anoxic zones that kill fish due to lack of oxygen, or even clog their gills. For an aquaculture producer, the sudden onset of a HAB can mean the total loss of production in a matter of days.
Until now, one of the greatest challenges has been the inability to predict the onset of a red tide far enough in advance to take preventive measures. However, a revolutionary study published in the prestigious journal Nature Communications by scientists from the University of Washington, the Georgia Institute of Technology, and the University of Rhode Island presents a new forecasting tool that could be a game-changer. Scientists have demonstrated that the oceanic microbiome undergoes a predictable and quantifiable shift just before a bloom begins, offering a crucial warning window.
The microbiome as a sentinel: A paradigm shift
Instead of focusing solely on the algae, the researchers proposed an innovative hypothesis: the free-living bacteria that coexist with phytoplankton are extremely sensitive to the subtlest chemical and physical changes in the environment. These bacteria quickly adjust their internal machinery—their proteins—to respond to these variations. The hypothesis was that this bacterial response, detectable at the peptide level (the components of proteins), occurs before the phytoplankton begins its exponential growth characteristic of a HAB.
The study focused on free-living bacteria, a key strategic decision. This not only facilitates the extraction of bacterial signals without interference from the abundant phytoplankton biomass, but also increases the likelihood that the identified biomarkers will be applicable to different bloom systems, rather than being tied to a particular algal species.
High-resolution molecular sspionage
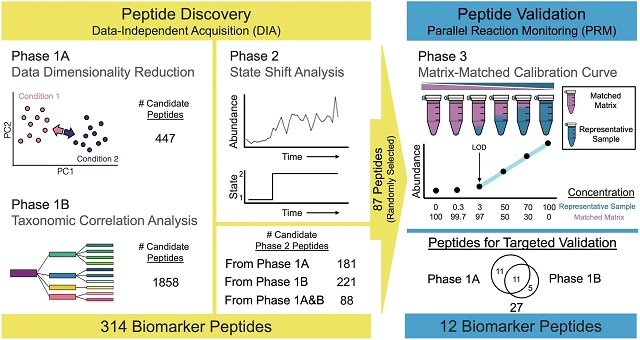
To test their hypothesis, the research team conducted high-resolution sampling in East Sound, Washington (USA), a dynamic coastal ecosystem. They collected bacterial microbiome samples every four hours during the complete cycle of two blooms of the diatom Chaetoceros socialis.
The experimental design was particularly robust:
- Discovery phase: They used a first pre-bloom period, characterized by more stable environmental parameters, to analyze thousands of bacterial peptides using advanced mass spectrometry techniques (metaproteomics) and to identify potential biomarker candidates.
- Validation phase: They tested these candidates in a second pre-bloom period that occurred 17 days later and under completely different nutrient conditions. The objective was to confirm if the same peptides showed the same predictive behavior despite the environmental variations.
This “discovery and validation” approach under different conditions lends great robustness to the findings, suggesting that the biomarkers are not a simple response to a local, specific condition.
12 Bacterial peptides for early warning
The analysis was a success. The team identified a final panel of 12 bacterial peptides that act as reliable predictive biomarkers.
The most important findings are:
Stay Always Informed
Join our communities to instantly receive the most important news, reports, and analysis from the aquaculture industry.
- Early warning confirmed: The 12 peptides showed a clear and definitive state change (described as “non-reverting”) at least 24 hours before the onset of the algal bloom in both studied events.
- Consistency and robustness: The pattern of these peptides was consistent across both blooms, even though the nutrient profiles (silicate, nitrate) in the water were very different in each case. This indicates that the biomarkers respond to a fundamental signal that precedes the bloom, regardless of certain environmental conditions.
- Origin of the biomarkers: The peptides came from common bacterial classes in marine ecosystems, such as Gammaproteobacteria, Flavobacteriia, and Verrucomicrobiae, which increases the probability that these findings can be generalized to other regions and types of blooms.
Interestingly, six of the twelve validated peptides were TonB-dependent receptors (TBDRs), a system that bacteria use to transport scarce resources like iron or carbohydrates. The fact that these metabolically expensive transport proteins are activated so early and in a coordinated manner suggests they are a highly sensitive indicator of the chemical changes that prepare the ground for a HAB.
Implications for aquaculture and coastal management
These discoveries open the door to a radical change in how the risks associated with HABs are managed in aquaculture.
- Anticipation and mitigation: A warning of more than 24 hours provides invaluable time for producers to take emergency measures: stop feeding to reduce fish stress, activate oxygenation systems, or even, if logistics permit, plan the relocation of cages.
- Overcoming current limitations: Unlike current monitoring methods, which often confirm a bloom when it is already underway and causing damage, this tool is truly predictive.
- Potential for an “On/Off” Test: The nature of the change detected in the peptides is so clear that it could facilitate the development of a rapid assay or test. Instead of complex and continuous monitoring, one could look for the “activation” of this signal to declare a pre-bloom alert, reducing development and operational costs.
Conclusion: Looking to the future
This study provides the first solid evidence that the analysis of the bacterial microbiome can generate reliable biomarkers to forecast the onset of harmful algal blooms. Although the authors themselves point out that more research is needed to validate these 12 peptides (and potentially others) in different ecosystems and with other types of algae—including highly toxic ones—the path is laid out.
The ability to anticipate a HAB, rather than just reacting to it, has long been a fundamental goal for science and the aquaculture industry. Thanks to these microscopic “sentinels,” that future is one step closer, promising safer, more sustainable, and more profitable management for global aquaculture.
Contact
Brook L. Nunn
Department of Genome Sciences, University of Washington
Seattle, WA, USA
Email: brookh@uw.edu
Reference (open access)
Mudge, M. C., Riffle, M., Chebli, G., Plubell, D. L., Rynearson, T. A., Noble, W. S., Kubanek, J., & Nunn, B. L. (2025). Harmful algal blooms are preceded by a predictable and quantifiable shift in the oceanic microbiome. Nature Communications, 16(1), 1-14. https://doi.org/10.1038/s41467-025-59250-y
Editor at the digital magazine AquaHoy. He holds a degree in Aquaculture Biology from the National University of Santa (UNS) and a Master’s degree in Science and Innovation Management from the Polytechnic University of Valencia, with postgraduate diplomas in Business Innovation and Innovation Management. He possesses extensive experience in the aquaculture and fisheries sector, having led the Fisheries Innovation Unit of the National Program for Innovation in Fisheries and Aquaculture (PNIPA). He has served as a senior consultant in technology watch, an innovation project formulator and advisor, and a lecturer at UNS. He is a member of the Peruvian College of Biologists and was recognized by the World Aquaculture Society (WAS) in 2016 for his contribution to aquaculture.