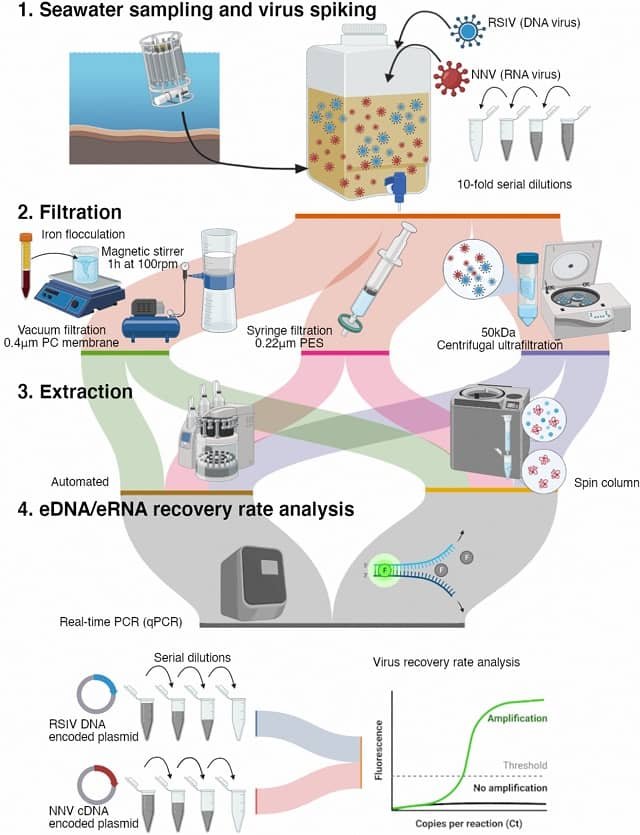
New York, USA.- An innovative tool that can confirm the recent presence of any given fish species in a sample of water is among the marvels to be highlighted at the first National Conference on Marine Environmental DNA, New York City, Nov. 29-30.
About 100 pioneering practitioners and users of eDNA science — a mighty complement to traditional marine life monitoring systems — will convene in Manhattan to detail and share discoveries, state-of-the-art technologies, and new methods.
A new tool created at The Rockefeller University, which will host the conference, offers, for example, a chemical shortcut for researchers to test for the eDNA of a specific, individual species in a water sample.
It makes use of the fact that every species leaves a trail of genetic residue — skin cells, excretions, other DNA — as it moves. Scientists can now test water and soil for these traces and identify which species left them behind.
The eDNA tester can confirm the genetic presence of a given species in a water sample within three days — a small fraction of the usual month or more involved in the current practice of lab testing for any and all species, or to mount an expedition with nets and analyze the results.
Its creator, Mark Stoeckle, Senior Research Associate at The Rockefeller University’s Program for the Human Environment (PHE), says many reasons make authorities want to know when a given marine species is present — to determine for example when to open or close a commercial fishery, or when dredging can be done without harm to marine life. New York Harbor, he notes, restricts dredging if winter flounder are present.
“Go Fish”
He likens his innovation to Go Fish, the children’s game in which a player asks another for a given rank of card, for example: do you have any jacks in your hand? Says Dr. Stoeckle. “In the case of New York, the question would be ‘Where in the harbor do we have winter flounder?'”
Stay Always Informed
Join our communities to instantly receive the most important news, reports, and analysis from the aquaculture industry.
The current cost to produce a Go Fish eDNA tool is $15 per sample (1 species); additional species can be added for $8 per sample.
Says conference lead organizer Jesse Ausubel, Director of the PHE: “‘Go Fish’ brings us close to a ‘chatbot’ or small smart personal assistant — like Siri, Alexa, or Cortana — that can quickly identify species from eDNA.”
“Presence of a species is easier to confirm than absence,” Dr. Stoeckle underlines. “Sampling may be conducted on the wrong day or at the wrong depth. Nevertheless, the genetic trail that animals leave behind is helping us find them without having them physically in hand — a breakthrough with major environmental and economic implications.”
National Conference on Marine Environmental DNA
Combined with traditional trawls with nets, tagging, visual observations, and acoustic instruments, experts believe remote eDNA sampling and analysis can one day help create near real-time monitoring of the marine environment.
In the US, several marine eDNA research hotspots have developed, including Monterey Bay, CA, New York / New Jersey, and Seattle, WA.
Expected at the conference are about 100 leading US scientists, officials, inventors, and investors exploring the emerging field of environmental DNA and its application in marine settings.
The event will highlight insights provided by eDNA to date and the potential of this new science to further enlarge our knowledge and inform ocean management.
Major themes:
Technology development — faster, cheaper, more portable
Bioinformatics — genetic reference databases, analytic software, data compatibility
eDNA biology — improving detection reliability, and relating eDNA abundance to species abundance
Organizers aim to initiate a commitment by leading scientists and stakeholders to take up eDNA as a cooperative national or regional research theme.
They also aim to encourage:
Federal, state, and local governments to incorporate eDNA into traditional ongoing . marine life surveys. (Monmouth University and Rockefeller U scientists are exploring integration of eDNA working with New Jersey Department of Marine Fisheries’ trawl surveys)
The private sector to collaborate in development of technologies to improve the speed and lower the cost of testing
Non-governmental organizations to help build genetic reference databases and monitor national and regional hotspots
Priority questions to be addressed include:
Whether and how the rate of decay of eDNA differs by taxa and context. Do some fish shed more than others? Do fish shed more than turtles? How do water temperature, sunlight, chemical variation, currents and turbulence, pressure, and other factors affect decay?
How to better calibrate the abundance of DNA in the water column as an index of abundance of specific species of fish and other animals
How to make eDNA reliable for very rare as well as more abundant species
How to formally integrate eDNA in the conduct of marine surveys, augmenting nets, cameras, and acoustic fish finders
What is needed to make eDNA data suitable for regulatory and policy purposes?
Says Paul Gaffney, Vice-Admiral (ret.), former President and Urban Coast Institute Ocean Policy Fellow at Monmouth University: “eDNA opens the door to cheap, frequent, widespread, potentially automated monitoring of the diversity, distribution, and abundance of aquatic life. Government agencies need to take notice.”
Bruce Nash, an innovator in adapting cutting-edge science for authentic student research, stressed the importance in years past of establishing protocols related to DNA barcoding, which identifies species from the DNA of tissue taken from physical specimens. Confirming a continuous chain of custody, time of testing, and other protocols made DNA barcode evidence sufficiently reliable to stand up in court.
To achieve reliable eDNA results, water or filtered material from the water needs to be stored and processed properly.
Dr. Nash will share the development of approachable and affordable methods that support eDNA work, including a new open-access tool developed at Cold Spring Harbor Laboratory’s DNA Learning Center for getting reliable identifications from the multitude of letters in the eDNA sequence data. Users upload their sequence data and then ride the Purple Line of DNA Subway in an appealing and intuitive interface to learn about the diversity contained in their sample.
Uses: eDNA’s applications to date include
Exploration
discovering species previously unknown in certain ranges
discovering rare species and others unknown to science (or absent from genome databases)
sampling remote, difficult-to-reach, and intriguing places
Assessment
health and stocks of fish in commercially harvested areas, informing decisions on when fisheries should open or close
range of marine animals
effect of protected area designation on fish and other marine animal populations and other forms of ecological restoration
effect of fish farming operations on native species
effect of offshore oil and gas operations or wind farms on marine life
effects of artificial reefs
effects of severe storms and other disturbances to marine ecosystems such as harmful algal blooms
Monitoring
presence of vulnerable, threatened or endangered species
presence of species dangerous to swimmers
impacts of climate change and variability
mapping marine animal diversity, distribution, migration and abundance, including invasive species, and species popular with sport fishers
Resolving mysteries (or not, in the case of Scotland’s Loch Ness monster)
Examples:
Sampling intriguing, remote, and difficult-to-reach intriguing places
Bob Ballard, who discovered the wreck of the Titanic in 1985, and colleagues on an expedition led by Dwight Coleman of the University of Rhode Island this month collected water from sediments from ancient sea caves including, for example, those about 70-100 meters deep on Osborn Bank in the Channel Islands, about 50 kilometers off the Southern California coast. The caves were above sea level 15,000 years ago and the team searched for ancient paleoshoreline features, and will analyse the samples for DNA that might have been left behind by human or other cave dwellers
Jesse Ausubel comments: “In cool, dark, undisturbed environments eDNA could persist long enough in water in the mud to provide clues about the critters, including humans, that lived in a spot thousands of years earlier.”
Scientist David Burg provided a set of fresh water samples from the Sea of Galilee and Jordan River to the Rockefeller U team, who found DNA of 15 fish species — more than half of known resident species. They include Galilee tilapia (Sarotherodon galilaeus) and blue tilapia (Oreochromis aureus), species that have sustained Sea of Galilee fishers for thousands of years to the present. In a 2006 book, marine biologist Eugene H. Kaplan speculated that one of those species was at the centre of St. Matthew’s New Testament story of the “miracle of the loaves and fishes” (depicted in a mosaic, dated to 420 A.D., in the Basilica of Sant’Apollinare Nuovo in Ravenna, Italy). There has been similar speculation that a Sea of Galilee tilapia was “St. Peter’s fish,” referred to in another Biblical passage.
An eDNA test this summer off Inkwell Beach on Martha’s Vineyard, MA discovered hard-to-spot leatherback turtles. Collection of eDNA at Inkwell Beach is part of the plot of Secrets from the Deep, latest of the popular Devlin Quick teenage detective books whose author, New York’s former sex crimes prosecutor Linda Fairstein, will attend the conference.
Last year, several dead leatherback washed ashore on the island, perhaps killed by boats. A GoFish eDNA turtle ‘dipstick’ might alert boaters to take extra care.
In June 2018, Dr. Stoeckle and colleague Jennifer Miksis-Olds sampled water from 500 meters depth to find elusive mid-water fish — species very easy to miss with nets on expensive trawls.
Tracking and mapping migrations of vulnerable, threatened, endangered and other species
In a major expedition this year, scientists led by Stanford Prof. Barbara Block at Hopkins Marine Station documented predators and prey in the White Shark Café — an area of the Pacific Ocean about half-way between Hawaii and the west coast of North America mysteriously visited regularly by white sharks. To determine what draws these predatory sharks to these seemingly inhospitable waters, they deployed a combination of satellite tags on white sharks, underwater robots, wind-powered Saildrones, and eDNA sampling to identify how and why animal species use these waters.
For three years, The Rockefeller University researchers have successfully monitored fish migrations in New York’s East and Hudson Rivers using only eDNA tests on weekly water samples. Over 30 months, the data snapshots created a moving picture of the presence or absence of several key fish species passing through, the results correlating closely with prior migration studies done with fishnet trawls.
Meanwhile, for 12 years New York City has been trying to coax the Alewife species of herring back to a former breeding habitat in the Bronx River, building fish ladders and reintroducing spawning adults. eDNA sampling of an 18-mile stretch of river this summer turned up no juveniles after the adults left to return the ocean, suggesting the need for further rehabilitation efforts.
Health and safety
Researchers have used eDNA to ‘sniff’ for sharks around beaches, and this year confirmed that a California boy’s shark bite was delivered by a Great White.
New York high school students drew 1 litre samples of water weekly from the fishing pier at Coney Island during the spring and summer of 2017 and discovered 34 species of marine life had passed by, including sharks and rays.
Invasive species
Foreign invader and pest species — both plant and animal — can be located and monitored quickly, easily and less expensively using eDNA instead of traditional methods. Examples of species already targeted in this way include lionfish in Bermuda, Asian black-spined toad and red-eared slider turtles in Australian waters, zebra mussels in the Great Lakes, and clams in the lakes of California and Nevada.
In Wisconsin, researchers documented five invasive species of marine zooplankton in the ballast water of ships plying Lake Superior, including the eDNA of a “bloody red shrimp” originally from the Black Sea area.
Encouraging citizen science
Students from Boynton Middle School are in a collaboration with Cornell University to find invasive fish species in their part of upstate New York.
And a Westchester high school senior spent the past year tracking water-loving mammals — river otter, muskrat, racoon, and beaver — by testing eDNA in streams in upstate New York and Rhode Island.
Limitations
Finding the eDNA of some species might not indicate its living presence in the vicinity. In their study of fish migration in the rivers surrounding Manhattan, for example, The Rockefeller University researchers found the DNA of species thought to have passed through humans and the wastewater treatment system — tilapia, salmon, red snapper — species you shouldn’t find swimming in the Hudson River. eDNA could therefore help identify vulnerable or threatened species being sold as food in local stores and restaurants.
Meanwhile, experts expect newer technologies will better detect the amount of DNA in a water sample but high concentrations might not indicate an abundance of animals passing through the water. It might be caused by an animal that is spawning, wounded or decaying, for example.
Says Alison Watts of the University of New Hampshire: “Modern genetic and acoustic tools provide complementary data identifying organisms at a range of distances, to comprehensively detect aquatic species. eDNA and passive acoustic monitoring are evolving technologies which may transform our understanding of marine communities.”
At the conference, Dr. Watts and co-author Jennifer Miksis-Olds will present a new paper (available at http://bit.ly/2zuyrqo): “The Ocean as a Living Sensor: Environmental DNA and Acoustics for Detecting Marine Life.”
Among many new eDNA-related technologies
Researchers working towards the automation and simplification of eDNA sampling are pursuing several interconnected technological directions. For example:
Using drones to collect water samples
Extracting eDNA from a water sample in the field (as it is easier to store DNA (a bit of goo on a filter) than the much larger water sample
Sequencing and analysing DNA in situ on board a sampling device, such as a remotely controlled glider, with digital results stored or relayed by satellite
Cold Spring Harbor “subway lines”
This is an innovation for analysing sequence data, with free and open access for all. It works for more than just eDNA sequences (microbiomes, etc.) but it works great for sequences eDNA’ers generate. (See https://dnasubway.cyverse.org/ and https://learning.cyverse.org/projects/dnasubway_guide/en/latest/step8.html )
At the University of Maryland, meanwhile, 3D printing is being deployed to create an ocean floor device that houses a water filter and pump that can collect eDNA samples at any depth.
California’s Monterey Bay Aquarium Research Institute is trying to integrate and automate collection of water, filtering of eDNA from water, and sequencing of the filtered DNA. Autonomous underwater vehicles and gliders could collect the water samples over large areas without sending humans out to sea.
Potential illustrations:
http://bit.ly/2FCqHZy
http://bit.ly/2P5R4qi
Background: eDNA
Almost 20 years ago, ecologist Pierre Taberlet of the Laboratoire d’Ecologie Alpine in France envisioned noninvasive sampling allowing genetic studies of free-ranging animals without the need to capture or even observe them.
After a decade of work, the technique emerged in papers such as: Species detection using environmental DNA from water samples, by Gentile Francesco Ficetola, Claude Miaud, Francois Pompanon, Pierre Taberlet, August 2008.
Taberlet’s fellow pioneers included Danish geneticist Eske Willerslev, who obtained ancient DNA directly from ice cores, and American marine biologist, Ann Bucklin, whose “Zoogene” project initiated in 2000 created a database of DNA type sequences for about 300 species of zooplankton.
Taberlet’s applications occurred mainly in freshwater habitats (paper here: http://bit.ly/2TNF0xM ). In the US, champions of the technique have included David Lodge of Cornell University, also mainly in a freshwater context. Lodge appreciated the charisma of eDNA in titles of his papers such as Conservation in a cup of water: estimating biodiversity and population abundance from environmental DNA, and ‘Sight unseen’, detection of rare aquatic species using environmental DNA.
In April, 2018, Taberlet co-authored with Aurelie Bonin, Lucie Zinger, and Eric Coissac the first book about eDNA: Environmental DNA For Biodiversity Research and Monitoring. The book aims to demonstrate the power and potential of environmental DNA as a research and conservation tool; describe available techniques and protocols; and guide researchers in efficient production of high-quality eDNA data and facilitate proper analysis and interpretation.
Marine eDNA 101: a Primer click here: http://bit.ly/2FDOBnC
Comments
“eDNA can turn millions of people into trustworthy environmental detectives.” – Linda Fairstein, New York lawyer and author of the Alexandra Cooper detective books
“Over 1,000 miles from shore we were able in 48 hours to identify the presence of white sharks in the water column beneath the ship using nanopore eDNA sequencing at sea. Censusing our oceans — knowing what is there or what we are losing — will be easier to document in the next decade with these powerful techniques.” – Barbara Block, Prothro Professor in Marine Sciences, Stanford University
“The Mid-Atlantic region is a leader in developing and deploying eDNA science and technology, and can benefit enormously because of the importance of marine fisheries and other living marine resources, and the need to minimize conflicts with navigation and proposed offshore wind farms that contribute to our Blue Economy.” – Tony MacDonald, Director, Urban Coast Institute, Monmouth University.
Source: The Rockefeller University
Editor at the digital magazine AquaHoy. He holds a degree in Aquaculture Biology from the National University of Santa (UNS) and a Master’s degree in Science and Innovation Management from the Polytechnic University of Valencia, with postgraduate diplomas in Business Innovation and Innovation Management. He possesses extensive experience in the aquaculture and fisheries sector, having led the Fisheries Innovation Unit of the National Program for Innovation in Fisheries and Aquaculture (PNIPA). He has served as a senior consultant in technology watch, an innovation project formulator and advisor, and a lecturer at UNS. He is a member of the Peruvian College of Biologists and was recognized by the World Aquaculture Society (WAS) in 2016 for his contribution to aquaculture.