Recirculating Aquaculture Systems (RAS) represent a fundamental advancement in the sustainable production of Atlantic salmon, enabling the cultivation of larger smolts in controlled, biosecure environments. However, the closed nature of these systems—their “Achilles’ heel”—can facilitate the accumulation and persistence of pathogens, turning a minor issue into a large-scale outbreak.
Traditionally, disease surveillance has relied on tissue sampling, an invasive and often lethal method that raises both economic and animal welfare concerns. This challenge prompts a key question: is it possible to detect the invisible enemies of salmon simply by analyzing the water they inhabit?
A recent study published by researchers from the Faroese Food and Veterinary Authority, the University of Copenhagen, and Hiddenfjord, conducted over 12 weeks at a commercial Atlantic salmon pre-smolt farm, evaluated the potential of environmental DNA and RNA (eDNA/eRNA) as an early-warning tool. The results not only validate this non-invasive technique but also unveil the complex dynamics of multiple infections within an RAS environment.
Key findings
- Environmental DNA/RNA (eDNA/eRNA) analysis of water is an effective, non-invasive tool for the early detection of salmon gill poxvirus (SGPV) and infectious salmon anaemia virus (ISAV-HPR0) in RAS.
- The viral load of SGPV in the water before fish stocking directly correlated with the severity and speed of the outbreak, acting as a valuable predictive indicator.
- A sequential infection pattern was observed: an initial SGPV outbreak was followed by secondary infections of ISAV-HPR0 and infectious pancreatic necrosis virus (IPNV).
- Pathogens can persist in the RAS environment between production cycles, highlighting the critical importance of disinfection to maintain system biosecurity.
A commercial RAS under the microscope
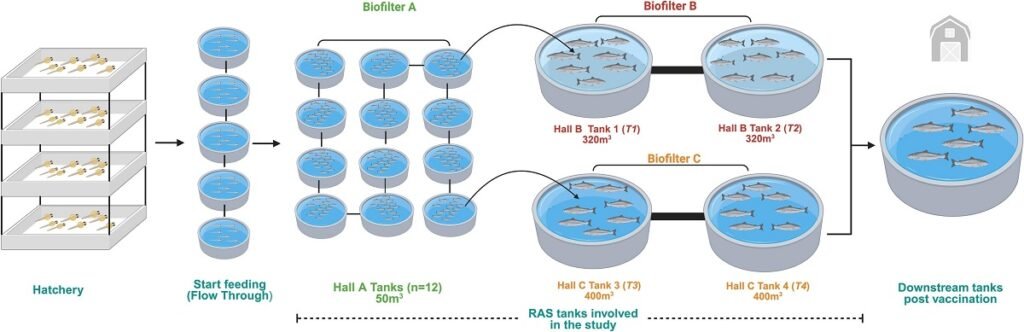
The study was conducted in a facility with three halls (A, B, and C), each operating as an independent RAS with its own biofilter. Naïve fish, with no prior exposure to pathogens, were reared in hall A and later transferred to halls B and C for grow-out.
Over twelve weeks, researchers monitored for the presence of five key pathogens known to persist in Faroese RAS:
- Salmon gill poxvirus (SGPV)
- Non-virulent infectious salmon anaemia virus (ISAV-HPR0)
- Infectious pancreatic necrosis virus (IPNV)
- Piscine orthoreovirus genotype 1 (PRV-1)
- The bacterium Flavobacterium psychrophilum
To do this, they compared results from gill and kidney swabs from the fish with those from water samples, which were filtered to capture the genetic material (eDNA/eRNA) shed by the fish and microorganisms into the environment.
Infection dynamics: One pathogen opens the door for others
The findings revealed a nearly identical sequential infection pattern in the two grow-out halls (B and C), demonstrating how one pathogen can weaken the fish and facilitate the entry of others.
SGPV: The primary trigger
The first pathogen to appear was SGPV, causing a clinical infection. The key difference between the two halls was the initial viral load in the water. Before the fish were introduced, water samples from Hall B already showed SGPV levels 100 times higher than those in Hall C.
This initial difference had drastic consequences:
Stay Always Informed
Join our communities to instantly receive the most important news, reports, and analysis from the aquaculture industry.
- In Hall B: The SGPV infection developed rapidly, peaking 9 days post-introduction and causing a sharp increase in mortality.
- In Hall C: With a low initial viral load, the infection progressed slowly, peaking at 23 days without causing a significant rise in mortality.
This result is crucial, as it proves that quantifying SGPV in water can function as a predictive indicator of the severity of a future outbreak.
A cascade of secondary infections
Following the peak of the SGPV infection, the fish became vulnerable to other pathogens. In hall B, the more affected hall, a concurrent increase in ISAV-HPR0 and IPNV was observed after 22 days. The opportunistic bacterium F. psychrophilum also showed small infection peaks following the SGPV outbreak in both halls.
This sequence suggests that the SGPV infection may suppress the fish’s immune system, creating a window of opportunity for secondary infections that complicate the clinical picture.
Water samples vs. swabs: How reliable is Environmental DNA?
The primary goal of the study was to validate eDNA/eRNA as an alternative to tissue sampling. The results varied depending on the pathogen, offering clear guidance on how to use this tool.
Strong correlation for gill pathogens
For the two viruses that primarily affect the gills, SGPV and ISAV-HPR0, a strong positive correlation was found between the viral load detected in gill swabs and that detected in water samples. This means that water analysis reliably and accurately reflects the infection status of the fish population. The reason is that infected epithelial cells from the gills are actively shed into the water, releasing the genetic material of both the virus and the fish, which can then be captured and analyzed.
Detecting internal pathogens and system persistence
For pathogens with predominantly internal infections, such as IPNV and PRV-1, no consistent correlation was found between kidney swabs and water samples. Interestingly, researchers often detected higher and more stable levels of these viruses in the water than in the fish.
This does not invalidate the technique but rather changes its interpretation: for internal pathogens, water analysis is excellent for confirming their presence and persistence in the RAS environment, but not necessarily for diagnosing the level of active infection in the fish. This reinforces the idea that pathogens can establish “house strains” that recirculate and persist in system components, such as biofilters.
Practical implications for salmon farming in RAS
This study transcends academia and offers tools directly applicable to producers:
- Predictive Monitoring: Regular water analysis to quantify SGPV can alert producers to a high risk of an outbreak even before fish show clinical signs.
- Proactive Management: Based on these findings, a tiered management approach is proposed. For example, if Cq values for SGPV in the water drop below a certain threshold (e.g., <25), monitoring should be intensified. If they continue to drop, mitigation strategies such as increasing water exchange, improving oxygenation, or halting feeding can be applied to reduce stress and slow disease progression.
- Improved Biosecurity: The confirmation that pathogens persist in the system underscores the critical need for rigorous disinfection protocols between production cycles to prevent contamination of new fish batches.
- Animal Welfare: By replacing lethal sampling, eDNA/eRNA monitoring reduces fish stress and mortality, aligning with the growing demands for welfare in aquaculture.
Ultimately, this study demonstrates that water analysis is not merely a scientific curiosity but a powerful surveillance tool that allows a shift from reactive to predictive and proactive management. Implementing monitoring programs based on eDNA/eRNA can significantly improve the biosecurity, fish health, and economic sustainability of salmon production in RAS.
Contact
Debes Hammershaimb Christiansen
National Reference Laboratory for Fish and Animal Diseases, Faroese Food and Veterinary Authority
Torshavn, Faroe Islands
Email: debesc@hfs.fo
Reference (open access)
Krishna, D., Petersen, P. E., Dahl, M. M., Egholm, I., Von Gersdorff Jørgensen, L., & Christiansen, D. H. (2025). Environmental DNA/RNA for non-invasive early detection and monitoring of pathogen dynamics in Atlantic salmon (Salmo salar) recirculating aquaculture systems (RAS). Aquaculture, 743060. https://doi.org/10.1016/j.aquaculture.2025.743060
Editor at the digital magazine AquaHoy. He holds a degree in Aquaculture Biology from the National University of Santa (UNS) and a Master’s degree in Science and Innovation Management from the Polytechnic University of Valencia, with postgraduate diplomas in Business Innovation and Innovation Management. He possesses extensive experience in the aquaculture and fisheries sector, having led the Fisheries Innovation Unit of the National Program for Innovation in Fisheries and Aquaculture (PNIPA). He has served as a senior consultant in technology watch, an innovation project formulator and advisor, and a lecturer at UNS. He is a member of the Peruvian College of Biologists and was recognized by the World Aquaculture Society (WAS) in 2016 for his contribution to aquaculture.