The Pacific white shrimp, Penaeus vannamei, has revolutionized the global aquaculture industry. Domesticated around 1980, this species has become the most farmed crustacean worldwide. However, the genetic implications of this rapid domestication remain a subject of ongoing scientific interest.
To better understand the genetic diversity of this species, researchers from CSIRO Agriculture and Food and Genics analyzed 44,522 single nucleotide polymorphisms (SNPs) from 21 globally distributed farmed populations and 86 wild P. vannamei. The results provide valuable insights into the genetic composition of this important aquaculture species.
- 1 A Complex Domestication History
- 2 Challenges in Managing Genetic Diversity
- 3 Recent Advances in Genotyping Tools
- 4 Wild Populations Show Weak Population Structure
- 5 Farmed Populations Show Low to Moderate Population Differentiation
- 6 Whole-Genome Sequencing Reveals Similar Nucleotide Diversity
- 7 Selection Sweeps Identify Outlier Regions and Genes
- 8 Conclusion
- 9 Entradas relacionadas:
A Complex Domestication History
The domestication of P. vannamei is a relatively recent effort dating back to the late 1980s. This process involved sourcing wild individuals from the eastern Pacific Ocean, primarily from regions ranging from Mexico to northern Peru.
Early domestication efforts focused on improving growth rates and disease resistance through selective breeding. As the industry expanded, the genetic diversity of farmed populations also increased, with genetic material exchanged between countries and regions.
Challenges in Managing Genetic Diversity
A key challenge in managing genetic diversity in P. vannamei breeding populations is the risk of inbreeding and reduced fitness due to the species’ high fecundity. Genotyping tools that inform genetic diversity levels and support more advanced breeding strategies are essential to sustain genetic gain and industry growth.
Recent Advances in Genotyping Tools
Recent advancements in genotyping tools have enabled the development of two SNP-based linkage maps and a 7K SNP genotyping array for P. vannamei. These tools have paved the way for genotype-to-phenotype studies and allowed the evaluation of genetic diversity in commercial breeding populations.
Wild Populations Show Weak Population Structure
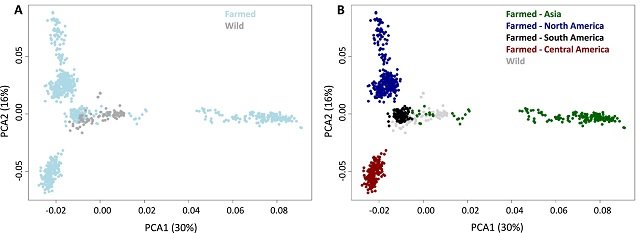
The study found that wild populations from Panama, Honduras, Guatemala, and Colombia exhibited weak population structure, with only 0–3% genetic differentiation. In contrast, wild populations from Mexico were more distinct, suggesting they may represent a separate subpopulation. This lack of strong population structure among wild populations may be attributed to the species’ ability to disperse and migrate across large distances.
Farmed Populations Show Low to Moderate Population Differentiation
When comparing farmed and wild populations, the study observed low to moderate population differentiation, with Asian farmed populations being the most divergent. This indicates that, while some genetic drift and selection have occurred in farmed populations, overall genetic diversity has been maintained.
Whole-Genome Sequencing Reveals Similar Nucleotide Diversity
To ensure unbiased results, the study utilized whole-genome sequencing to identify 4.13 million SNPs in a subset of farmed and wild shrimp.
Stay Always Informed
Join our communities to instantly receive the most important news, reports, and analysis from the aquaculture industry.
The results showed that observed nucleotide diversity levels in wild and farmed animals were similar, suggesting that the domestication process has not significantly reduced genetic diversity. This is likely due to the repeated introduction of wild shrimp into farmed populations, helping maintain the species’ genetic diversity.
Selection Sweeps Identify Outlier Regions and Genes
The study also identified a small collection of outlier regions and 20 protein-coding genes that may have been subject to selection sweeps. These regions and genes may have been favored by artificial selection in farmed populations, leading to changes in the species’ phenotype.
Conclusion
The study provides a comprehensive snapshot of the genetic diversity and divergence within the Pacific white shrimp, P. vannamei. The findings can be summarized as follows:
- Domestication has led to distinct genetic clusters: Farmed shrimp populations from different geographic regions have undergone significant allele frequency changes since domestication in the 1980s, resulting in distinct clusters.
- Little genetic diversity has been lost in farmed populations: Despite domestication and selection for performance traits, the genetic diversity of farmed shrimp has remained relatively high, with minimal loss compared to wild populations.
- Wild shrimp possess unique genetic traits: Whole-genome sequencing data revealed that wild shrimp have an excess of private alleles not found in farmed populations. This suggests that wild shrimp possess a unique genetic composition that could benefit aquaculture.
- Introduction of wild broodstock can complement genetic diversity: The study suggests that incorporating wild-caught broodstock into farmed populations could help complement genetic diversity, especially in areas where it has been reduced.
These findings have important practical implications for the management and conservation of P. vannamei populations. By incorporating wild broodstock into farmed populations, the aquaculture industry can help maintain genetic diversity and potentially improve the resilience and adaptability of farmed shrimp populations.
Contact
James Kijas
CSIRO Agriculture and Food
Brisbane, Queensland 4067, Australia
Email: james.kijas@csiro.au
Reference (open access)
Kijas, J., Carvalheiro, R., Menzies, M., McWilliam, S., Coman, G., Foote, A., Moser, R., Franz, L., & Sellars, M. (2025). Genome-wide SNP variation reveals genetic structure and high levels of diversity in a global survey of wild and farmed Pacific white shrimp. Aquaculture, 597, 741911. https://doi.org/10.1016/j.aquaculture.2024.741911
Editor at the digital magazine AquaHoy. He holds a degree in Aquaculture Biology from the National University of Santa (UNS) and a Master’s degree in Science and Innovation Management from the Polytechnic University of Valencia, with postgraduate diplomas in Business Innovation and Innovation Management. He possesses extensive experience in the aquaculture and fisheries sector, having led the Fisheries Innovation Unit of the National Program for Innovation in Fisheries and Aquaculture (PNIPA). He has served as a senior consultant in technology watch, an innovation project formulator and advisor, and a lecturer at UNS. He is a member of the Peruvian College of Biologists and was recognized by the World Aquaculture Society (WAS) in 2016 for his contribution to aquaculture.