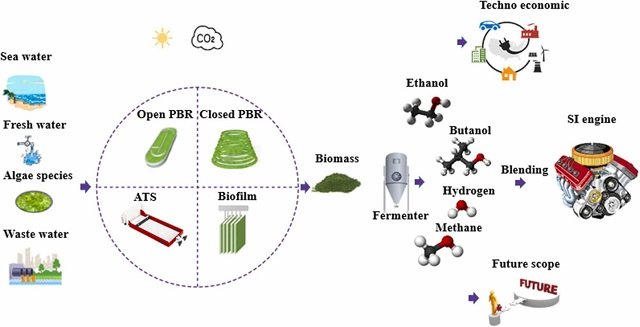
The PhycoCosm data portal allows anyone to freely investigate the genes of more than 100 algae.
The Science
Interested in the genomes of algae? You now have one place where you can browse the genetic blueprints of these photosynthetic organisms. PhycoCosm is one of the largest data repositories of its kind, with an interactive browser that allows algal scientists and enthusiasts to look deep into more than 100 algal genomes, compare them, and visualize supporting experimental data.
The Impact
Algae are important organisms. They play key roles in global carbon cycling, are sources of natural products, and have potential as biofuel feedstocks. Understanding the genetic underpinnings responsible for these traits takes us closer to harnessing algae for bioenergy and the greater bioeconomy. By bringing together publicly available genomic data on algae into one place, PhycoCosm allows users to easily compile data that answers what genes are present in which organisms, when are they expressed, and what they do.
Summary
The name PhycoCosm stems from the Greek phykos, meaning seaweed. Seaweeds are algae, but many kinds of algae exist. During the course of evolution, algal organisms sprung up in almost all branches of eukaryotes (organisms with a nucleus). In part because of this evolutionary diversity, algal genomes can be incredibly complex and difficult to sequence and understand.
A team of researchers led by Algal Genomics Program lead Igor Grigoriev and data scientist Alan Kuo at the U.S. Department of Energy (DOE) Joint Genome Institute (JGI), a DOE Office of Science User Facility located at Lawrence Berkeley National Laboratory (Berkeley Lab), have unveiled PhycoCosm in the Nucleic Acids Research journal. The genome portal reinforces the JGI’s new strategic focus on exploring algal biology, diversity, and ecology.
Stay Always Informed
Join our communities to instantly receive the most important news, reports, and analysis from the aquaculture industry.
PhycoCosm has many components that make it useful for scientific inquiry: it shows a full phylogenetic tree of more than 100 sequenced algal genomes — many sequenced by the JGI — so researchers can easily explore the evolutionary and functional relationships among the different algae. Scientists can use PhycoCosm’s genome browser to see their favorite alga’s predicted genes and their organization, and analyze genes in the nucleus and organelles such as the chloroplast. In the cases where data are available, a user can also analyze gene expression from published experiments; study DNA methylation, which gives clues to how an organism may tune gene expression; and investigate corresponding proteins. PhycoCosm’s built-in connection to the DOE Systems Biology Knowledgebase (KBase) also allows researchers to study how metabolites flow through and transform in algae.
PhycoCosm joins other online data portals and tools created by JGI researchers, including the Integrated Microbial Genomes and Microbiomes (IMG/M) system for microbial and metagenome datasets, MycoCosm for comparative analysis of fungal genomes, and Phytozome for comparing plant genomes.
Kuo and Grigoriev encourage scientists to both utilize PhycoCosm’s data repository and contribute to it. And for those still needing their algae sequenced, Grigoriev and Kuo recommend they apply through the JGI’s Community Science Program.
Contacts:
BER Contact
Ramana Madupu, Ph.D.
Program Manager
Biological Systems Sciences Division
Office of Biological and Environmental Research
Office of Science
US Department of Energy
Ramana.Madupu@science.doe.gov
PI Contact
Igor Grigoriev, Ph.D.
Algal Program Lead
DOE Joint Genome Institute
ivgrigoriev@lbl.gov
Publication (open access):
Grigoriev et al. PhycoCosm, a comparative algal genomics resource. Nucleic Acids Research. 2020 October. doi: 1093/nar/gkaa898
Editor at the digital magazine AquaHoy. He holds a degree in Aquaculture Biology from the National University of Santa (UNS) and a Master’s degree in Science and Innovation Management from the Polytechnic University of Valencia, with postgraduate diplomas in Business Innovation and Innovation Management. He possesses extensive experience in the aquaculture and fisheries sector, having led the Fisheries Innovation Unit of the National Program for Innovation in Fisheries and Aquaculture (PNIPA). He has served as a senior consultant in technology watch, an innovation project formulator and advisor, and a lecturer at UNS. He is a member of the Peruvian College of Biologists and was recognized by the World Aquaculture Society (WAS) in 2016 for his contribution to aquaculture.