The field of aquaculture is on the verge of a breakthrough! Single-cell genomics, a rapidly advancing technology, is transforming the way we understand the biology of fish, mollusks, crustaceans, and other aquatic species. This exciting approach allows scientists to delve deeper than ever before and unveil the unique molecular profiles of individual cells.
For decades, researchers have relied on “bulk” methods to understand the inner workings of living organisms. These methods analyze a mixed set of cells, averaging out the complexities they contain. But what if we could listen to the unique voice of each cell?
Single-cell genomics, a new and powerful set of techniques, is poised to revolutionize our understanding of organisms cultivated in aquatic environments. In this regard, scientists from The Roslin Institute at The University of Edinburgh published a scientific review that provides a useful source of knowledge for researchers seeking to apply single-cell genomics to address the biological challenges facing the global aquaculture sector through a better understanding of cellular biology.
Individual cells, a significant impact
Traditionally, researchers relied on the study of whole tissues, providing an average picture of all the cells they contain. Single-cell genomics breaks this limitation. By analyzing individual cells, we can uncover hidden diversity within tissues. Different types of cells, with specialized functions, work together to create a complex biological system. Single-cell genomics sheds light on these intricate interactions and provides a much richer understanding of how organisms function.
Single-cell genomics allows scientists to analyze the unique molecular footprint of individual cells. This provides a much richer picture compared to general methods, revealing cellular activity that keeps fish healthy and thriving.
The study published in the scientific journal Reviews in Aquaculture begins with a brief overview of single-cell transcriptomics, followed by descriptions of possible applications and impacts that single-cell genomics can bring to aquaculture research.
Additionally, scientists explore the barriers to achieving such impacts, along with possible solutions and considerations when designing/executing experiments relevant to wet lab work and also about subsequent data analysis and interpretation.
The future of aquaculture lies in cells
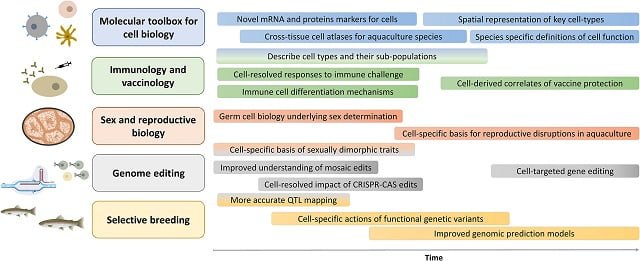
The article has provided a high-level overview. Let’s delve into some of the specific applications of single-cell genomics in aquaculture research:
- Creation of cellular atlases: Single-cell data can be used to create detailed “cellular atlases” mapping the various types of cells present in different tissues of an organism. This information provides a fundamental understanding of cellular biology and serves as a valuable resource for future research.
- Revolutionizing immunology: By identifying and characterizing immune cell types, single-cell genomics can accelerate vaccine development and enhance our understanding of how farmed fish combat diseases.
- Understanding host-pathogen interactions: This technology enables scientists to study how pathogens interact with host cells at the single-cell level, providing insights into disease mechanisms and paving the way for more targeted interventions.
- Genome editing: Single-cell genomics can be combined with genome editing techniques to identify gene function and improve the efficiency of breeding programs.
- Uncovering secrets of sex and reproduction: By analyzing the transcriptome of reproductive cells, scientists can gain valuable insights into sex determination, gonadal development, and ultimately improve reproductive success in captivity.
- Revolutionizing selective breeding: Single-cell data can help identify specific genes underlying important traits, leading to more efficient and specific selective breeding programs.
Challenges and solutions: Taking a step forward
While single-cell genomics offers a treasure trove of information, there are some hurdles to overcome:
- Unique challenges for aquatic species: Many commercially important aquaculture species are “ectothermic,” meaning their body temperature depends on the environment. This presents special challenges in obtaining high-quality cells for analysis.
- The genomic gap: Compared to humans and model organisms, the genomes of many aquaculture species are not fully characterized. This requires additional expertise in bioinformatics to effectively analyze the data.
The good news is that researchers are actively developing solutions! Techniques are being refined to work with delicate aquatic cells, and bioinformatics tools are constantly evolving to handle less characterized genomes.
Conclusion
“Single-cell genomics is rapidly being adopted in aquaculture species and, when applied alongside other emerging technologies, will revolutionize our understanding of the specific cellular basis for traits important for sustainability and production goals,” conclude the study’s authors.
Thus, single-cell genomics is poised to revolutionize aquaculture. By uncovering hidden secrets within individual cells, we can create a future with healthier fish, better disease management, and sustainable aquaculture practices. As technology matures and challenges are addressed, the potential for transformative advances in aquaculture is immense.
The study was funded by the Scottish Universities Life Sciences Alliance, the Data Driven Innovation Initiative of the University of Edinburgh, the Biotechnology and Biological Sciences Research Council, and the Norwegian Seafood Research Fund.
Contact
Daniel J. Macqueen
The Roslin Institute and Royal (Dick) School of Veterinary Studies, The University of Edinburgh, Midlothian, UK.
Email: daniel.macqueen@roslin.ed.ac.uk
Reference (open access)
Daniels, R. R., Taylor, R. S., Robledo, D., & Macqueen, D. J. (2023). Single cell genomics as a transformative approach for aquaculture research and innovation. Reviews in Aquaculture, 15(4), 1618-1637. https://doi.org/10.1111/raq.12806
Editor at the digital magazine AquaHoy. He holds a degree in Aquaculture Biology from the National University of Santa (UNS) and a Master’s degree in Science and Innovation Management from the Polytechnic University of Valencia, with postgraduate diplomas in Business Innovation and Innovation Management. He possesses extensive experience in the aquaculture and fisheries sector, having led the Fisheries Innovation Unit of the National Program for Innovation in Fisheries and Aquaculture (PNIPA). He has served as a senior consultant in technology watch, an innovation project formulator and advisor, and a lecturer at UNS. He is a member of the Peruvian College of Biologists and was recognized by the World Aquaculture Society (WAS) in 2016 for his contribution to aquaculture.
